Pavankumar Videm
Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Topic: Single Cell
- Learning Pathway: Applying single-cell RNA-seq analysis
- Learning Pathway: Epigenetics data analysis with Galaxy
- Learning Pathway: Introduction to Galaxy and Single Cell RNA Sequence analysis
- Learning Pathway: Applying single-cell RNA-seq analysis in Coding Environments
Tutorials
- Statistics and machine learning / Unsupervised Analysis of Bone Marrow Cells with Flexynesis 🧐
- Transcriptomics / Small Non-coding RNA Clustering using BlockClust ✍️ 🧐
- Transcriptomics / Reference-based RNA-Seq data analysis ✍️ 🧐
- Transcriptomics / Genome-wide alternative splicing analysis 📝 🧐
- Transcriptomics / RNA-RNA interactome data analysis ✍️
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana ✍️ 🧐
- Imaging / Content Tracking and Verification in Galaxy Workflows with ISCC-SUM 🧐
- Proteomics / MaxQuant and MSstats for the analysis of label-free data 🧐
- Proteomics / Metaproteomics tutorial 🧐
- Proteomics / Single Cell Proteomics data analysis with bioconductor-scp 📝 🧐
- Contributing to the Galaxy Training Material / Creating content in Markdown 🧐
- Epigenetics / DNA Methylation data analysis 📝
- Epigenetics / CUT&RUN data analysis ✍️ 🧐
- Epigenetics / Formation of the Super-Structures on the Inactive X ✍️ 🧐
- Single Cell / Analysis of plant scRNA-Seq Data with Scanpy ✍️
- Single Cell / Filter, plot and explore single-cell RNA-seq data with Scanpy (Python) 🧐
- Single Cell / Importing files from public atlases 🧐
- Single Cell / Pre-processing of 10X Single-Cell ATAC-seq Datasets ✍️ 🧐
- Single Cell / Single-cell ATAC-seq standard processing with SnapATAC2 📝 ⚙️ 🧐
- Single Cell / Downstream Single-cell RNA analysis with RaceID ⚙️
- Single Cell / Matrix Exchange Format to ESet | Creating a single-cell RNA-seq reference dataset for deconvolution 🧐
- Single Cell / Filter, plot and explore single-cell RNA-seq data with Scanpy 🖥 🧐
- Single Cell / Inferring single cell trajectories with Scanpy 📝 🧐
- Single Cell / Bulk matrix to ESet | Creating the bulk RNA-seq dataset for deconvolution 🧐
- Single Cell / Pseudobulk Analysis with Decoupler and EdgeR 📝 🧐
- Single Cell / Converting between common single cell data formats ⚙️ 🧐
- Single Cell / Inferring single cell trajectories with Monocle3 🧐
- Single Cell / Single-cell quality control with scater ⚙️
- Single Cell / Clustering 3K PBMCs with Seurat 📝 🧐
- Single Cell / Removing the effects of the cell cycle ⚙️
- Single Cell / Filter, plot, and explore single cell RNA-seq data with Seurat 📝 🧐
- Single Cell / Generating a single cell matrix using Alevin and combining datasets (bash + R) 📝 ⚙️ 🧐
- Single Cell / Combining single cell datasets after pre-processing 🧐
- Single Cell / Inferring single cell trajectories with Monocle3 (R) 📝 🧐
- Single Cell / Inferring single cell trajectories with Scanpy (Python) 🧐
- Single Cell / Filter, plot, and explore single cell RNA-seq data with Seurat (R) 📝 🧐
- Single Cell / Pre-processing of 10X Single-Cell RNA Datasets ✍️ 🧐
- Single Cell / Converting NCBI Data to the AnnData Format 📝 ⚙️ 🧐
- Single Cell / Generating a single cell matrix using Alevin 🧐
- Single Cell / Clustering 3K PBMCs with Scanpy ✍️ 🧐
- Ecology / Production d'indicateurs champs de bloc 🧐
Slides
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana ✍️ 🧐
- Single Cell / Single-cell Formats and Resources 🧐
FAQs
Video Recordings
- Single Cell / Pre-processing of 10X Single-Cell RNA Datasets 💬 🗣
- Single Cell / Clustering 3K PBMCs with Scanpy 💬 🗣
Events
- Workshop on high-throughput sequencing data analysis with Galaxy 🧑🏫
- A practical introduction to bioinformatics and RNA-seq using Galaxy 🧑🏫
- Workshop on high-throughput sequencing data analysis with Galaxy 🧑🏫
- Galaxy Training Academy 2025 🧑🏫
- From Data to Discovery: Metagenomics, RNA-Seq - NGS Bioinformatics with Galaxy 🧑🏫
- From data to discovery - Galaxy workshop at University of Graz 🧑🏫
- LOVE DATA week with intro to Galaxy & GTN 🎪
- Galaxy Training Academy 2024 🧑🏫
GitHub Activity
github Issues Reported
54 Merged Pull Requests
See all of the github Pull Requests and github Commits by Pavankumar Videm.
-
 Update dev topic with links to matrix and SIG
dev
Update dev topic with links to matrix and SIG
dev -
 Fix glossary heading duplication in tutorial.md
imaging
Fix glossary heading duplication in tutorial.md
imaging -
 Update single-cell editors and add publication
single-cell
Update single-cell editors and add publication
single-cell -
 Differential isoform analysis - add a warning and tips on annotation …
transcriptomics
Differential isoform analysis - add a warning and tips on annotation …
transcriptomics -
 minor updates to imaging tutorials
imaging
minor updates to imaging tutorials
imaging
Reviewed 42 PRs
We love our community reviewing each other's work!
-
 Create a new tutorial on the ISCC-SUM suite
template-and-toolsimaging
Create a new tutorial on the ISCC-SUM suite
template-and-toolsimaging -
 added workshop in Graz
template-and-tools
added workshop in Graz
template-and-tools -
 MaxQuant and MSstats for the analysis of label-free data - update maxquant version and results
proteomics
MaxQuant and MSstats for the analysis of label-free data - update maxquant version and results
proteomics -
 flexynesis GTN - unsupervised task
template-and-toolsstatistics
flexynesis GTN - unsupervised task
template-and-toolsstatistics -
 Improve tutorial text for clarity of the video recording
single-cell
Improve tutorial text for clarity of the video recording
single-cell
News
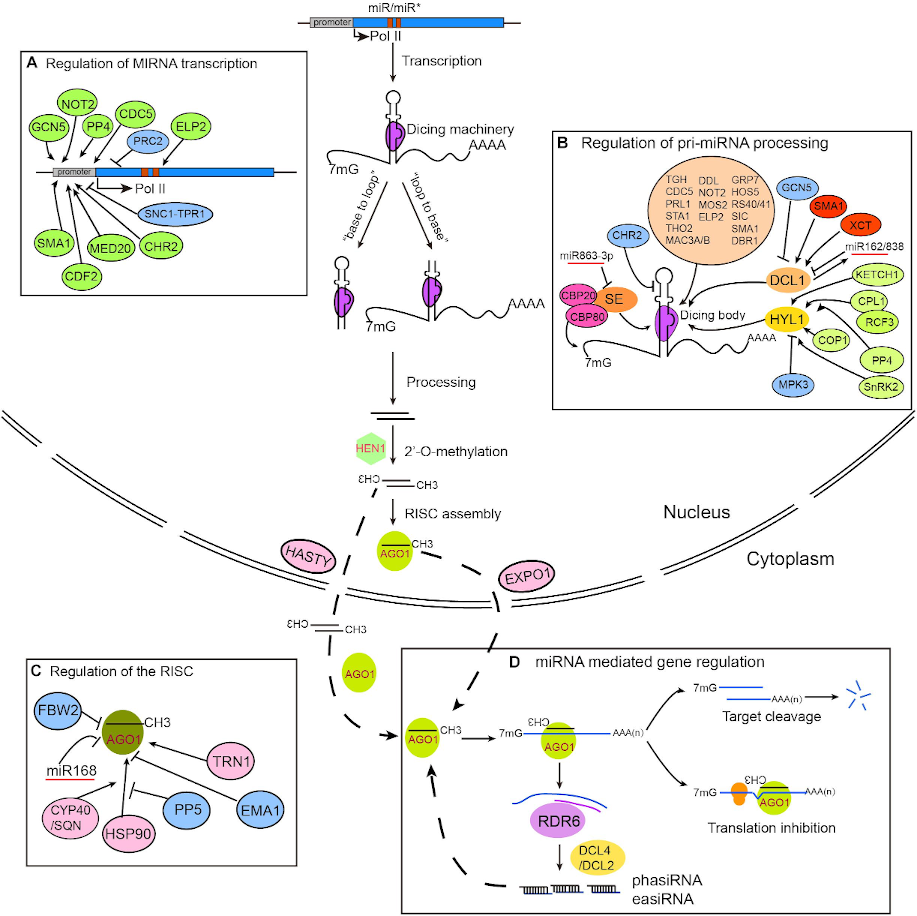
New Tutorials: Whole transcriptome analysis of Arabidopsis thaliana
10 April 2021
Plant lovers are in luck! A new tutorial on this fascinating kingdom has been added to the GTN. It details the necessary steps to identify potential targets of brassinosteroid-induced miRNAs. Brassinosteroids are phytohormones that have the ability to stimulate plant growth and confer resistance against abiotic and biotic stresses. These characteristics have led to great interest in the field of biotechnology due to its potential for increasing agricultural productivity.
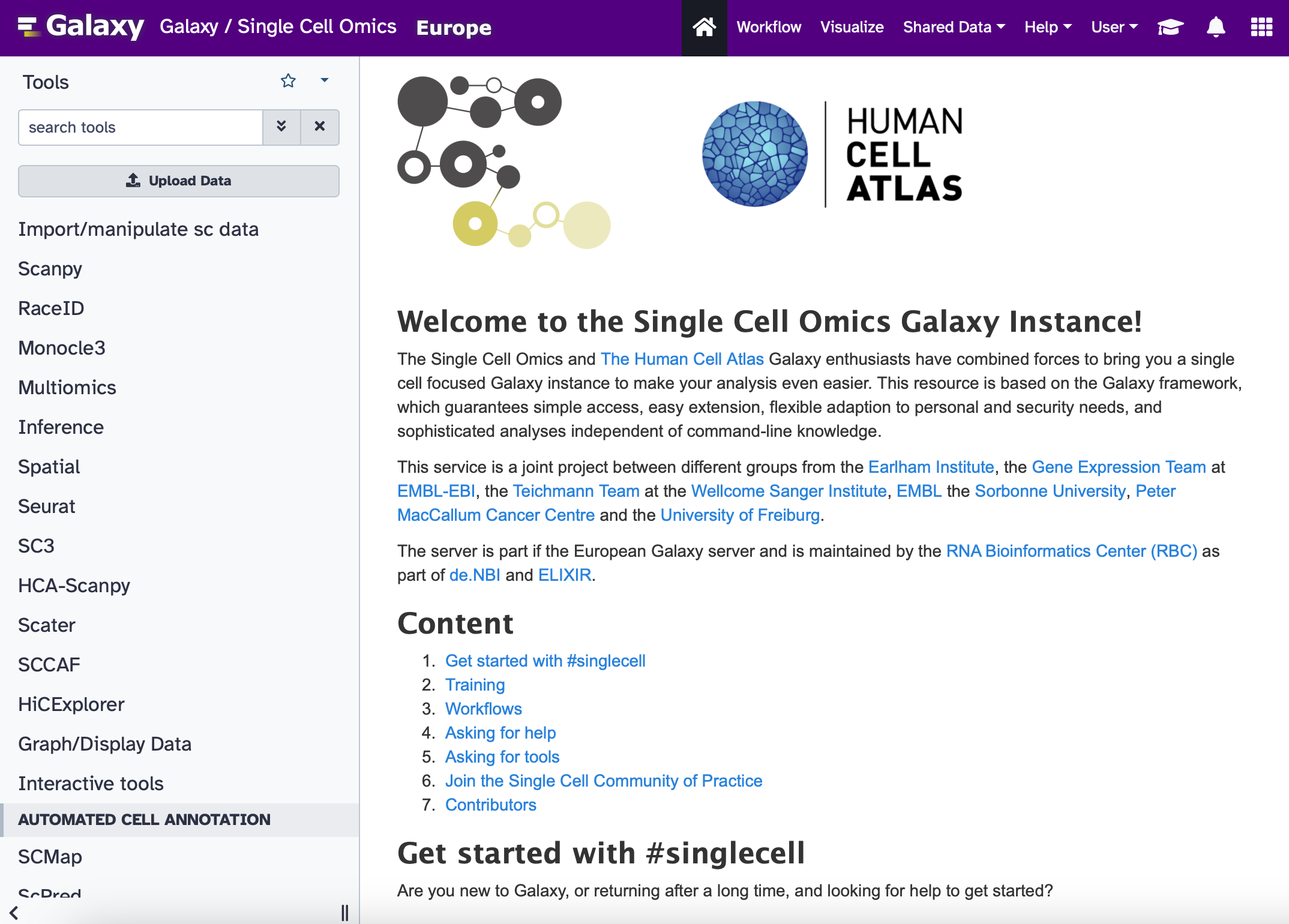
Single cell subdomain re-launch: Unified and feedback-driven
12 October 2023
The European Galaxy Days 2023 CoFest combined the forces of administrator, developer and trainer to update and re-launch the single cell Galaxy instance. Where previously there were two subdomains each with their own sets of tools, there is now a unified subdomain with re-categorized tools that makes sense for users.

Galaxy Single-cell Community: Year in Review
22 December 2023
🚀Embarking on a cosmic journey, the Galaxy Single-cell Community has clustered together to unveil a constellation of tools, making strides in RNA-stellar discoveries and creating out-of-this-world workflows. With a commitment to battling work duplication across the multiverse, this community is boldly charting a course for global domination, proving that when it comes to bioinformatics, the Galaxy is the limit!✨
