Secretome Prediction
Under Development!
This tutorial is not in its final state. The content may change a lot in the next months. Because of this status, it is also not listed in the topic pages.
| Author(s) |
|
| Reviewers |
|
OverviewQuestions:
Objectives:
How to predict cellular protein localization based upon GO-terms?
How to combine multiple localization predictions?
Requirements:
Predict proteins in the cellular secretome by using GO-terms.
Predict proteins in the cellular secretome by using WolfPSORT.
Combine the results of both predictions.
Time estimation: 30 minutesLevel: Intermediate IntermediateSupporting Materials:Published: Apr 13, 2017Last modification: Apr 8, 2025License: Tutorial Content is licensed under Creative Commons Attribution 4.0 International License. The GTN Framework is licensed under MITpurl PURL: https://gxy.io/GTN:T00235version Revision: 15
Secretome Prediction using GO annotations and localization prediction
Introduction
The cellular secretome contains both proteins that are secreted by cells and proteins that are shed from the cellular surface. Here, we describe an approach to predict those proteins in an input list that would be expected in the cellular secretome. This approach combines Gene Ontology (GO) annotation and the WoLF PSORT algorithm for localization prediction.
We chose to include all proteins that are annotated as, or predicted to be, lysosomal proteins. Lysosomal proteins are routinely secreted by malignant and non-malignant cells in high amounts, due to “leakiness” of the mannose-6-phosphate receptor pathway [1,2]. Furthermore, we chose to exclude proteins annotated as being part of extracellular organelles, e.g. exosomes. While exosomes are secreted by malignant and non-malignant cells, exosomal proteins are expected in the secretome at very low amounts, if not especially enriched for.
For secretome prediction, we combine localization data from the Gene Ontology database with a classical protein localization prediction algorithm (WoLF PSORT). The workflow was designed for sensitivity, i.e. a protein predicted by at least one of the used tools will be included in the output. To change this, follow the instructions in the box Comment: Customizing the Workflow below.
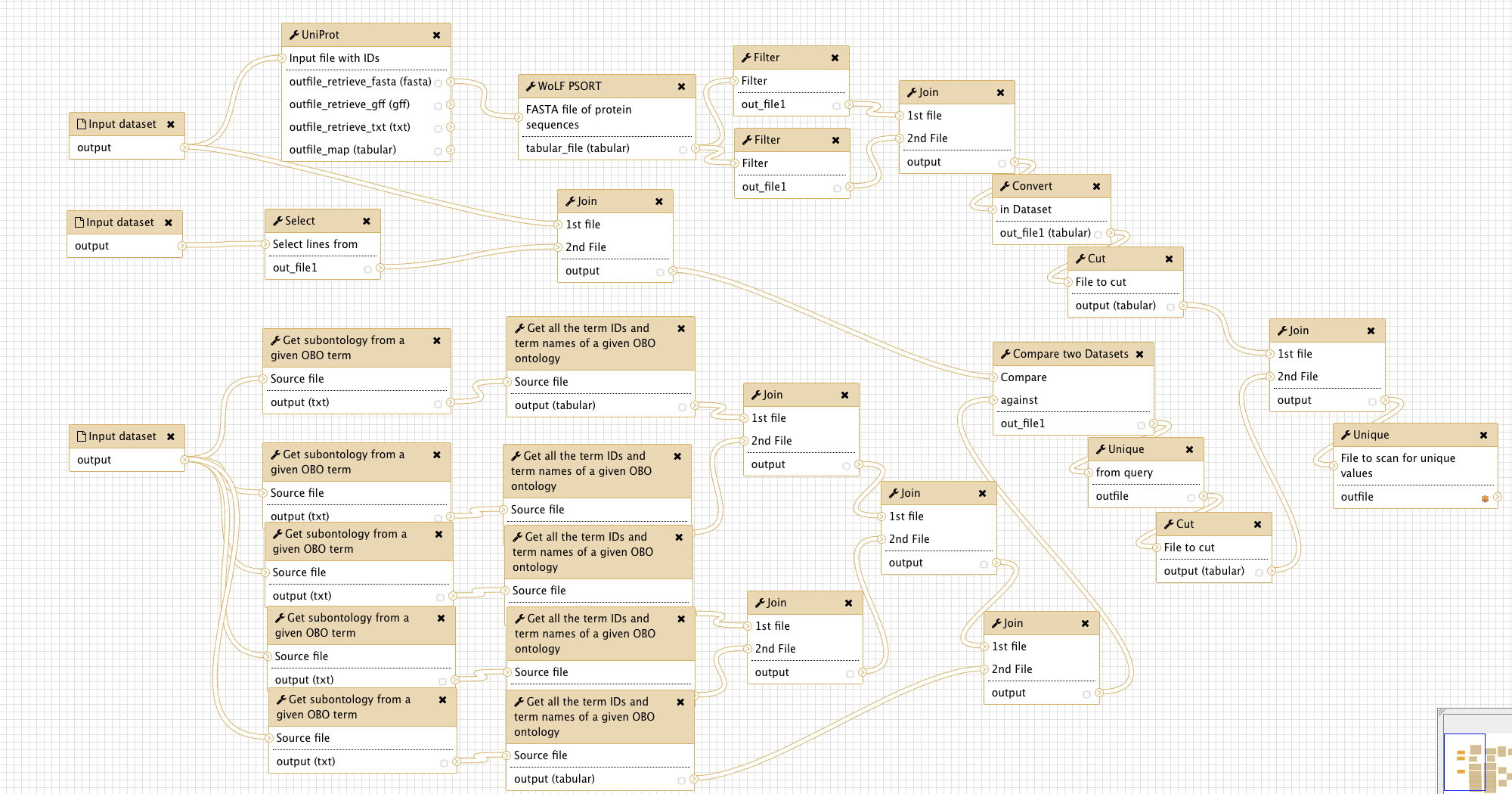
Overview
The figure below gives an overview of the Galaxy workflow:
Input
The workflow needs three input files:
-
A tabular file, the first column containing uniprot accession numbers of the proteins of interest. Test data
Comment: Test dataThe provided test dataset for input 1 is a list of human proteins, identified by LC-MS/MS in the cellular supernatant of MDA-MB-231 cells. The dataset was originally published in (Sigloch et al., BBA, 2016).
-
The complete uniprot GO database for the organism of interest, available via FTP. To download the human GOA file needed for the test input, paste the following link to the Galaxy upload tool: ‘ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/HUMAN/goa_human.gaf.gz’
Comment: Uniprot Gene Ontology Association (GOA) files -
The complete GO Open Biomedical Ontology (OBO), i.e. “GO term tree”, accessible at http://purl.obolibrary.org/obo/go/go.obo
Comment: Customizing the WorkflowThis workflow was designed for sensitivity, not for specificity. If you need to increase the specificity, you have the following possibilities, with decreasing efficiency:
- Switch the setting
Output lines appearing inof the last Join tool (last tool before the final Unique tool fromAll lines [-a 1 -a 2]toBoth 1st & 2nd file. Thus, your output will contain only those proteins that are equally predicted by both methods used.- (Only after doing 1.) Adding another way of localization prediction, i.e. another database or another prediction algorithm.
- Replacing WoLF PSORT tool by a more precise localization prediction tool. If you choose this approach, remember that you will probably have to adjust the settings for all tools in the workflow that are processing the WoLF PSORT tool output.
Citation
If you use this workflow directly, or any derivative of it, in work leading to a scientific publication, please cite:
F.C. Sigloch, J.D. Knopf, J. Weißer, A. Gomez-Auli, M.L. Biniossek, A. Petrera, et al., Proteomic analysis of silenced cathepsin B expression suggests non-proteolytic cathepsin B functionality, Biochim. Biophys. Acta - Mol. Cell Res. 1863 (2016) 2700–2709. doi:10.1016/j.bbamcr.2016.08.005. https://www.ncbi.nlm.nih.gov/pubmed/27526672
Literature
[1] B. Schröder, C. Wrocklage, A. Hasilik, P. Saftig, The proteome of lysosomes., Proteomics. 10 (2010) 4053–76. doi:10.1002/pmic.201000196.
[2] J. Reiser, B. Adair, T. Reinheckel, Specialized roles for cysteine cathepsins in health and disease, J. Clin. Invest. 120 (2010) 3421–3431. doi:10.1172/JCI42918.