| Author(s) |
|
Posted on: 30 March 2021 purlPURL: https://gxy.io/GTN:N00009
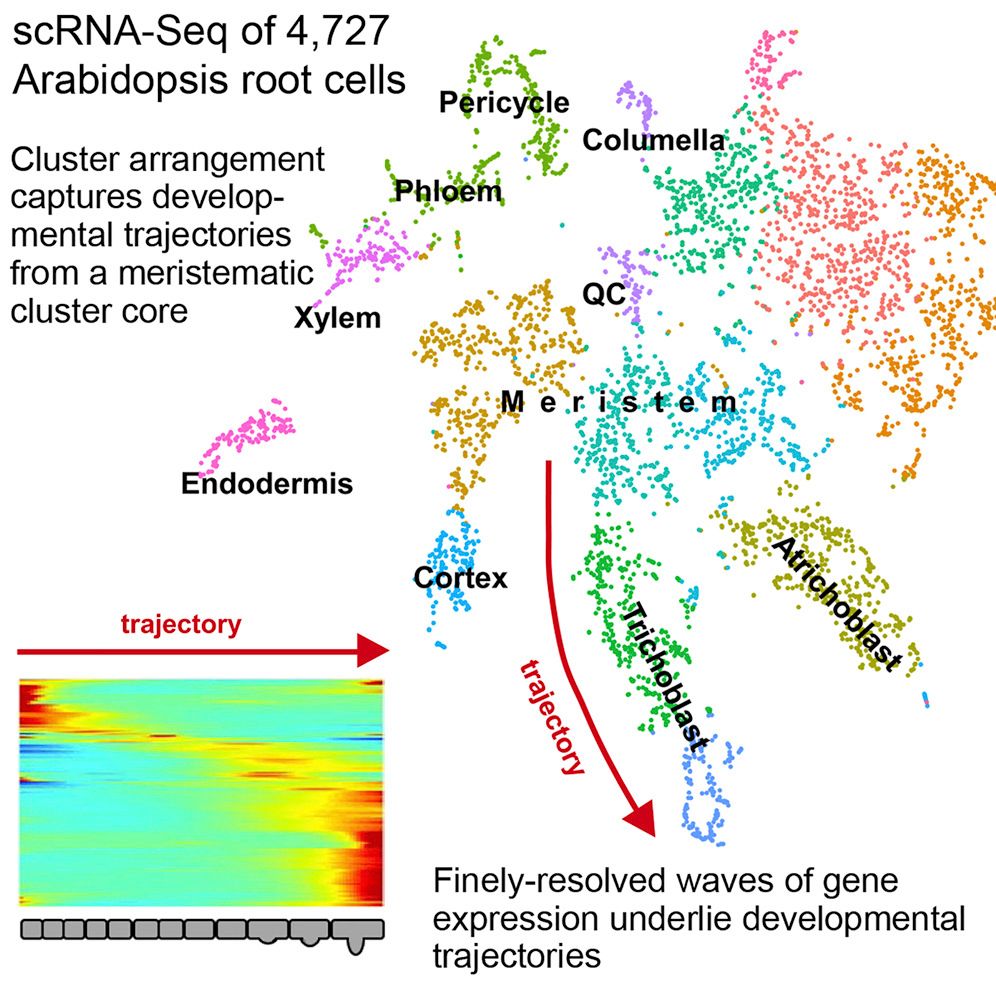
Single cell RNA-seq analysis is a cornerstone of developmental research and provides a great level of detail in understanding the underlying dynamic processes within tissues. In the context of plants, this highlights some of the key differentiation pathways that root cells undergo.
This tutorial replicates the paper Denyer et al. 2019, where the major plant cell types are recovered in the data as well as distinguishing between QC and meristematic cells.
View MaterialReferences
- Denyer, T., X. Ma, S. Klesen, E. Scacchi, K. Nieselt et al., 2019 Spatiotemporal Developmental Trajectories in the Arabidopsis Root Revealed Using High-Throughput Single-Cell RNA Sequencing. Developmental Cell 48: 840–852.e5. 10.1016/j.devcel.2019.02.022