Quantification of single-molecule RNA fluorescence in situ hybridization (smFISH) in yeast cell lines
| Author(s) |
|
| Reviewers |
|
OverviewQuestions:
Objectives:
How do I analyze fluorescence markers in a 2D image?
How do I create labels from the detected bright spots?
How can I count the number of detected spots automatically?
Requirements:
Perform a 2D spots/blobs detection in an image fetched from IDR
Count the detected spots and blobs
- Introduction to Galaxy Analyses
- tutorial Hands-on: FAIR Bioimage Metadata
- tutorial Hands-on: REMBI - Recommended Metadata for Biological Images – metadata guidelines for bioimaging data
- tutorial Hands-on: Introduction to Image Analysis using Galaxy
Time estimation: 30 minutesLevel: Introductory IntroductorySupporting Materials:Published: Mar 20, 2025Last modification: Nov 26, 2025License: Tutorial Content is licensed under Creative Commons Attribution 4.0 International License. The GTN Framework is licensed under MITpurl PURL: https://gxy.io/GTN:T00498version Revision: 5
The objective is to detect multiple bright spots in an image using a basic computer vision and image processing technique for identification and localization of regions of high intensity within an image. These spots often correspond to features of interest, such as fluorescent markers in biological imaging. Such an approach can be beneficial in single-cell/single-molecule imaging experiments, such as RNA single-molecule fluorescence in situ hybridization (smFISH), as these experiments can resolve the spatial and temporal distribution of individual RNA molecules with high resolution.
In this tutorial, we will try to identify RNA molecules in yeast cell lines. However, such an approach can be re-used for the identification of any fluorescence spots in biological images!
AgendaIn this tutorial, we will deal with:
Get data from IDR
The testing dataset for this tutorial can be obtained from the Image Data Repository (IDR). This dataset was published by Li et al. (2019) in Scientific Data.
In this publication, the authors used smFISH to quantify the kinetic expression of STL1 and CTT1 mRNAs in single Saccharomyces cerevisiae cells upon NaCl osmotic stress.
Let’s fetch this FAIR dataset using a Galaxy Tool! We will download the channel 5-TMR and selected the 10th z-stack for analysis.
Hands On: Download images from the IDR
If you are logged in, create a new history for this tutorial.
To create a new history simply click the new-history icon at the top of the history panel:
- IDR Download ( Galaxy version 0.45) with the following parameters:
- “How would you like to specify the IDs of images to download?”:
As text (comma-separated list of IDs or a valid IDR link)- “Image IDs to download”:
4496770- *“Which Images do you want to download”:
Exported TIFF (single channel, single stack)- “Name of the channel to download”:
5-TMR- “z-plane of images to download”:
10- “Image frame to download”:
0- “Limit the download to a selected region of the image?”:
No, download the entire image plane- “Skip failed retrievals?”:
No- “Download images in a tarball?”:
No- Extract dataset with the following parameters:
- param-collection “Input List”:
Download IDR/OMERO(output of IDR Download tool)- “How should a dataset be selected?”:
The first dataset- Rename galaxy-pencil the generated file to
RNA_input.tiff.
Improve image contrast
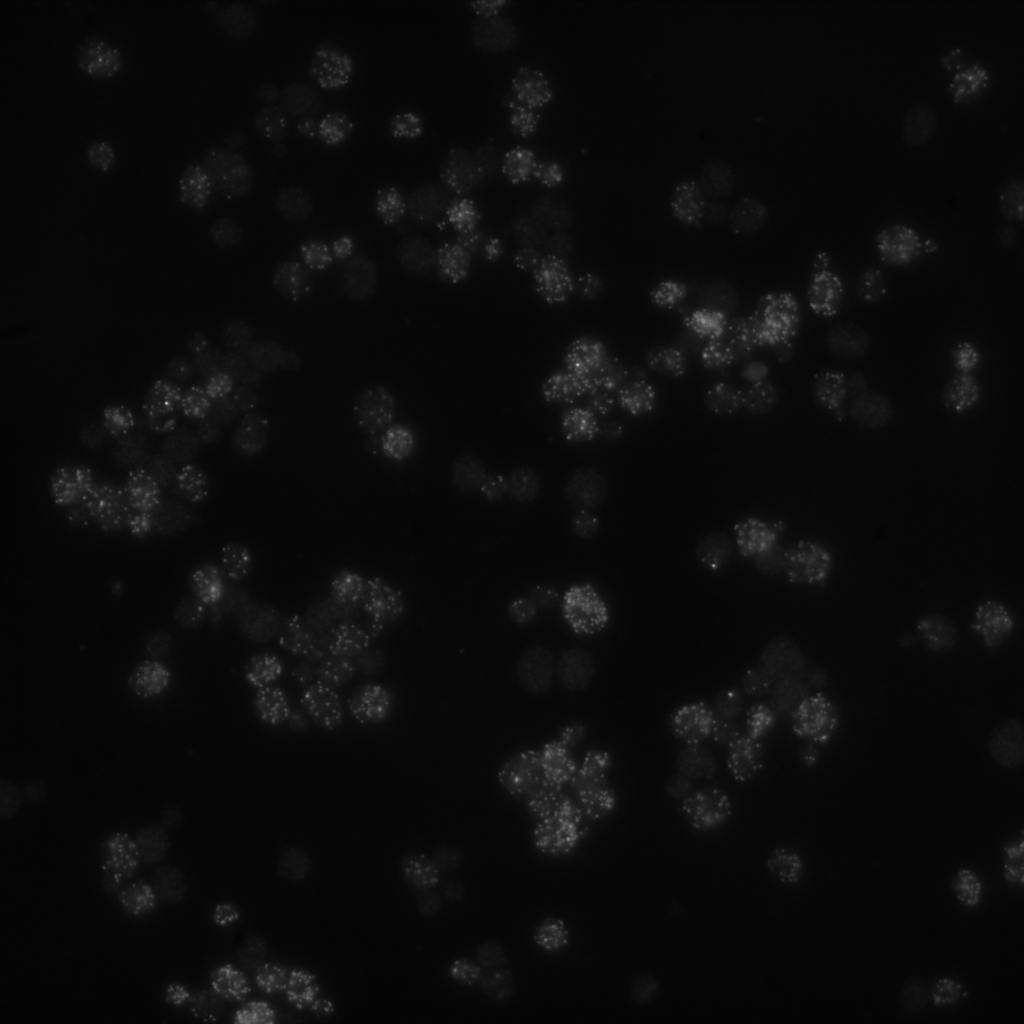
When inspecting the image visually, we get the impression that the image is entirely black:
It’s not the best quality for a first visual inspection!
As described in the Galaxy Imaging Introduction Tutorial, the original image is 16-bit and the intensity values are spread over a very small fraction of the range of intensity values that can be represented using 16 bits.
Therefore, for improved visibility the intensity histogram of the image can be normalized first.
We will normalize the histogram to improve the contrast. We do this using a Contrast Limited Adaptive Histogram Equalization (CLAHE) approach.
Hands On: Normalize Histogram
- Perform histogram equalization ( Galaxy version 0.18.1+galaxy0) with the following parameters to normalize the histogram of the image:
- param-file “Input image”:
RNA_input.tifffile- “Histogram equalization algorithm”:
CLAHE- Rename galaxy-pencil the generated file to
RNA_input_normalized.tiff.- Click on the visualise icon galaxy-visualise of the file to visually inspect the image using the Tiff Viewer visualization plugin.
Your image should now look a bit brighter:
We can now clearly make out the presence of many RNA spots!
RNA spots detection
We can now try to detect blobs and measure their intensity… Let’s run a specific tool for this!
Hands On: Perform 2-D spot detection
- Perform 2-D spot detection ( Galaxy version 0.1+galaxy0) with the following parameters to detect the 2D spots:
- param-file “Intensity image or a stack of images”:
RNA_input_normalized.tifffile- “Starting time point (1 for the first frame of the stack) “:
1- “Ending time point (0 for the last frame of the stack)”:
0- “Detection filter”:
Laplacian of Gaussian- “Minimum scale”:
1- “Maximum scale”:
2- “Minimum filter response (absolute)”:
0.25- “Minimum filter response (relative)”:
0- “Image boundary”:
10- Rename galaxy-pencil the generated file to
RNA_spot_detected.
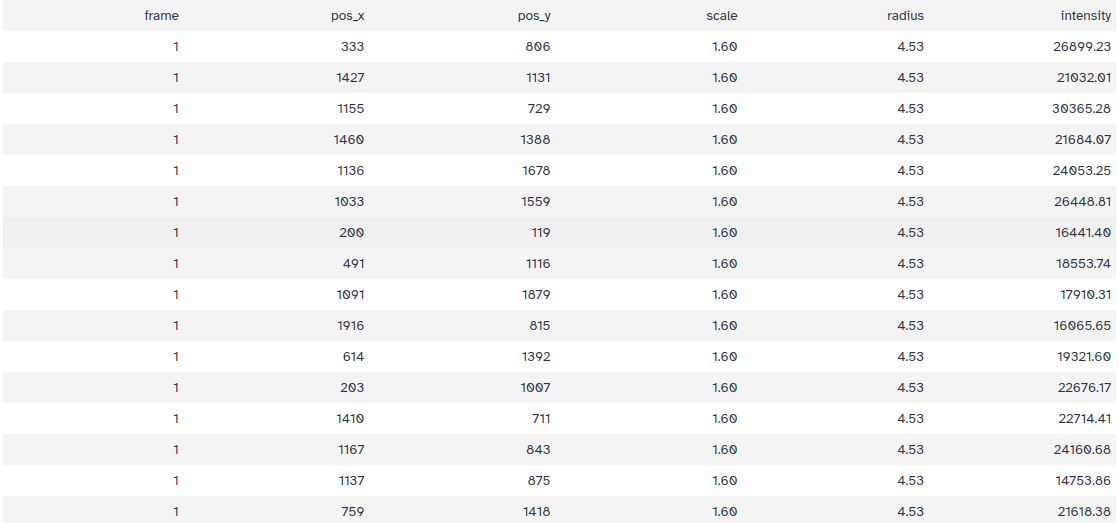
The tool produces a TSV file containing all detections, with the following columns:
- frame: The frame of the image stack
- pos_x: The horizontal coordinate of the detection
- pos_y: The vertical coordinate of the detection
- scale: The scale at which the detection was found
- radius: The radius of the detected spot
- intensity: The mean intensity of the spot
The TSV output will have 1649 lines, meaning 1649 spots detected!
Convert spots coordinates to labels
Let’s try now to visualize our results! The first step is to create a label map from the TSV file… This can be nicely done by converting the point coordinates to a label map and we have a tool for it!
Please notice that the width and height of the image are necessary inputs to corretly overlay the label map in the next step! You can get these info by running the “Show image info” tool on the original image!
Hands On: Create the label map from the points coordinates
- Convert point coordinates to label map ( Galaxy version 0.4+galaxy0) with the following parameters to detect the 2D spots:
- param-file “Tabular list of points”:
RNA_spot_detected.tsvfile- “Width of output image”:
2048- “Height of output image “:
2048- “Tabular list of points has header”:
Yes- “Swap X and Y coordinates”:
No- “Produce binary image”:
No- Rename galaxy-pencil the generated file to
RNA_labels.tiff.- Click on the visualise icon galaxy-visualise of the file to visually inspect the image using the Tiff Viewer visualization plugin.
A label image will be created… If you want to visualize the labels, you can run again the normalize histogram step:
but this is not necessary for the next and final step: overlay the label to the original image!
Overlay labels to the original image
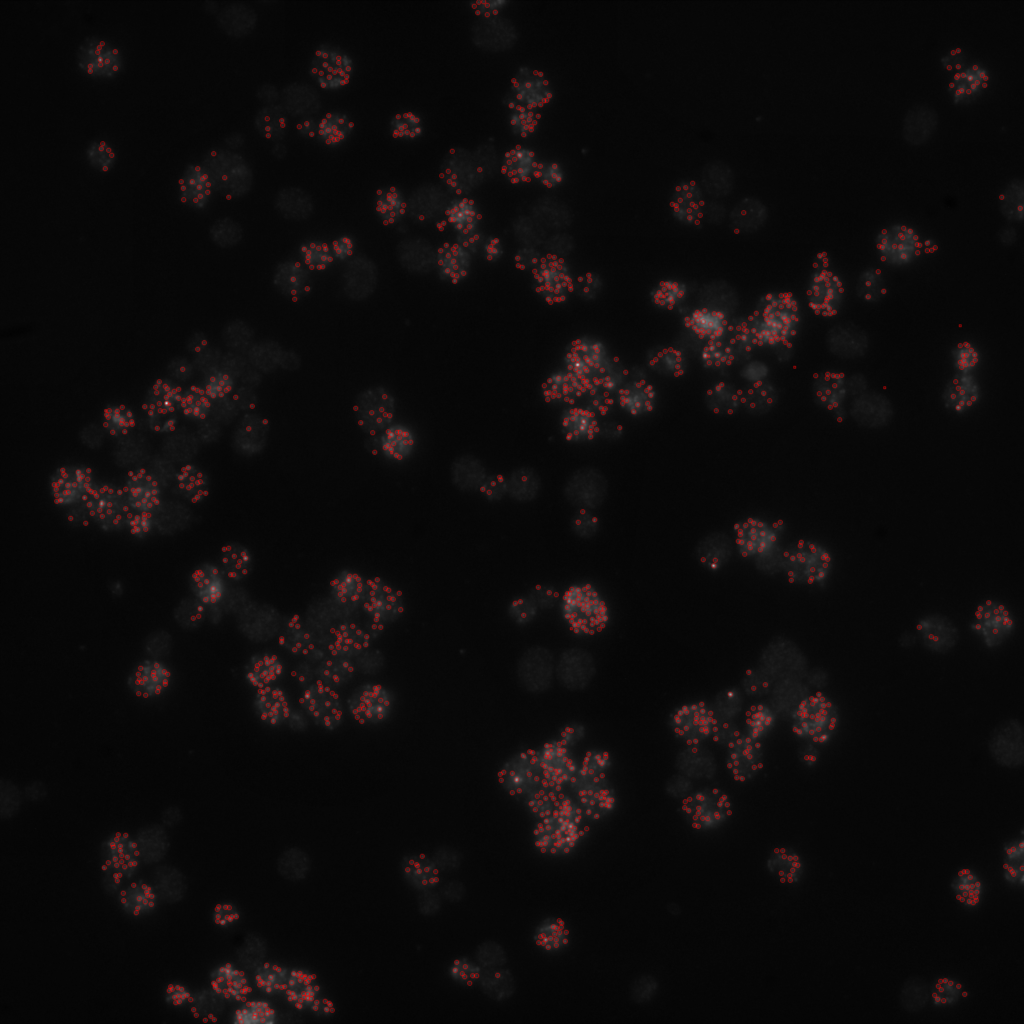
Results can be overlayed with the original image.
Hands On: Segment image
- Overlay images ( Galaxy version 0.0.4+galaxy4) with the following parameters to convert the image to PNG:
- “Type of the overlay”:
Segmentation contours over image- param-file “Intensity image”:
RNA_input_normalized.tifffile- param-file “Label map”:
RNA_labels.tifffile (output of Convert binary image to label map ( Galaxy version 0.5+galaxy0))- “Contour thickness”:
1- “Contour color”:
red- “Show labels”:
No- “Label color”:
yellow
Final result should look like this:
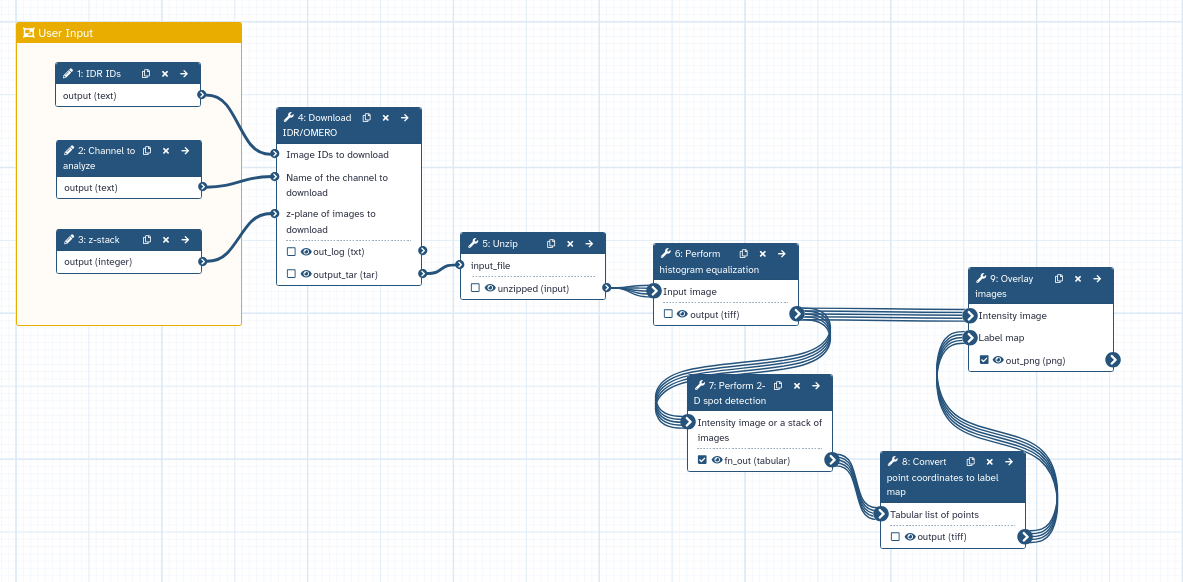
Create 2D spot detection workflow
The full workflow can now be created!
Since you are probably going to run it on several images fetched from IDR we suggest changing the setting of the IDR Download tool by downloading imaging in a tarball:
Hands On: Create the workflow for 2D spot detection
- Name it “2D_spot_detection”.
- Change “Download images in a tarball?”:
Yesin the IDR Download tool tool- Include the Unzip ( Galaxy version 6.0+galaxy0)
- Connect the output of param-file 4: Download IDR/OMERO to the “input_file” input of tool 5: Unzip.
- Connect the output of param-file 4: Unzip to the “Input Image” input of tool 6: Perform histogram equalization.
- Edit the workflow you just created:
- Select “Input dataset” from the list of tools. The step param-file 1: Input Dataset appears.
- Select “Text Input” from the list of tools. The step param-file 2: Text Input appears.
- Select “Integer Input” from the list of tools. The step param-file 3: Integer Input appears.
- Change the “Label” of param-file 1: Input Dataset to
IDR IDs.- Change the “Label” of param-file 2: Text Input to
Channel selection.- Change the “Label” of param-file 3: Integer Input to
z-stack selection.- Connect the output of param-file 1: Input Dataset to the “Image IDs to download” input of tool 4: Download IDR/OMERO.
- Connect the output of param-file 2: Text Input to the “Name of the channel to download” input of tool 4: Download IDR/OMERO.
- Connect the output of param-file 3: Integer Input to the “z-plane of images to download” input of tool 4: Download IDR/OMERO.
- Mark the results of tool 7: Perform 2-D spot detection and tool 9: Overlay images as the primary outputs of the workflow (by clicking on the checkboxes of the outputs).
The final workflow should look like this:
Conclusion
In this exercise, you imported images into Galaxy from IDR, detect RNA single molecules using the 2D spot detection tool and visualize them on the original intensity image. Finally, you created a reusable workflow which can be used on a tarball of IDR images.