Fine-tuning Protein Language Model
Contributors
| Author(s) |
|
Questions
How to load large protein AI models?
How to fine-tune such models on downstream tasks such as post-translational site prediction?
Objectives
Learn to load and use large protein models from HuggingFace
Learn to fine-tune them on specific tasks such as predicting dephosphorylation sites
Requirements
- slides Slides: Basics of machine learning
- tutorial Hands-on: Basics of machine learning
- slides Slides: Classification in Machine Learning
- tutorial Hands-on: Classification in Machine Learning
last_modification Published: Mar 21, 2025
last_modification Last Updated: Apr 8, 2025
Language Models (LM)
- Powerful LMs “understand” language like humans
- LMs are trained to understand and generate human language
- Popular models: GPT-3, Llama2, Gemini, …
- Trained on vast datasets with billions of parameters
- Self-supervised learning
- Masked language modeling
- Next word/sentence prediction
- Can we train such language models on protein/DNA/RNA sequences?
- LM for life sciences - DNABert, ProtBert, ProtT5, CodonBert, RNA-FM, ESM/2, BioGPT, …
- Many are available on HuggingFace
Bidirectional Encoder Representations from Transformers (BERT)
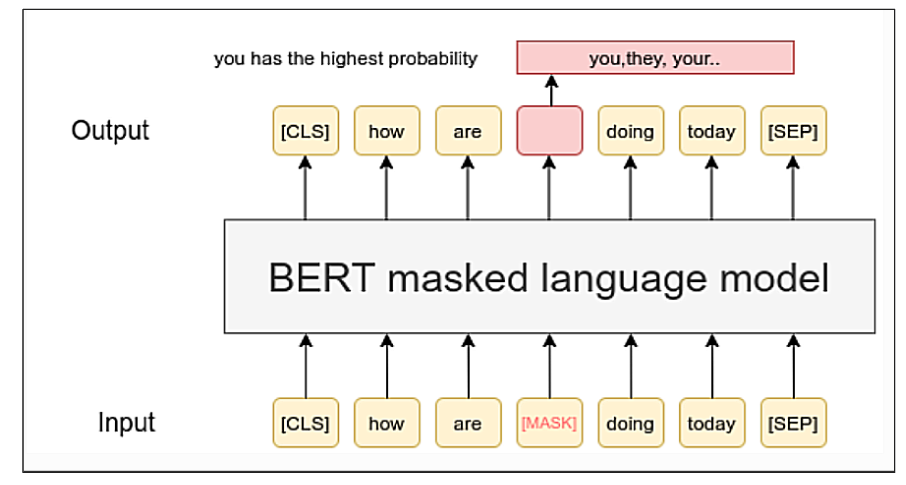
Language Models (LMs)
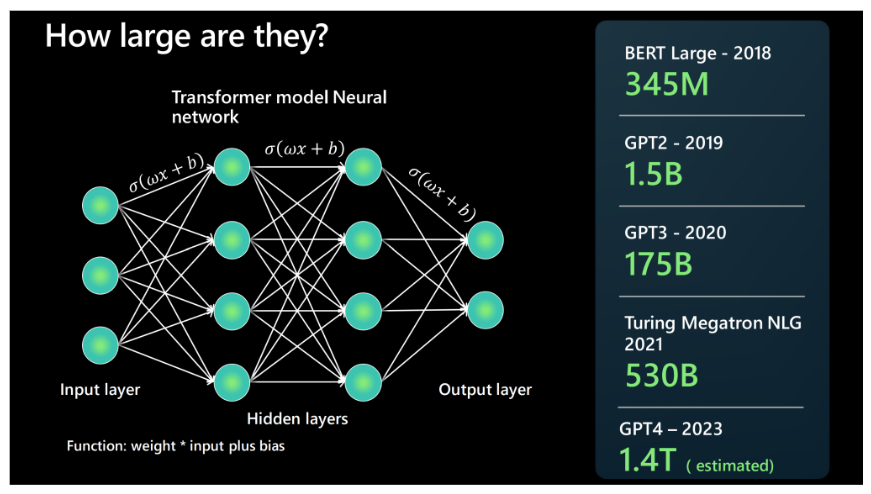
Protein Language Model (pLM)
- Models trained on large protein databases
- Big fantastic database (> 2.4 billions sequences), Uniprot, …
- Popular architectures such as BERT, Albert, T5, T5-XXL …
- Key challenges in training such models
- Large number of GPUs needed for training: (Prot)TXL needs > 5000 GPUs
- Expertise needed in large scale AI training
- Most labs and researchers don’t have access such resources
- Solution: fine-tune pre-trained models on downstream tasks such as protein family classification
- Benefits: requires smaller data, less expertise, training time and compute resources
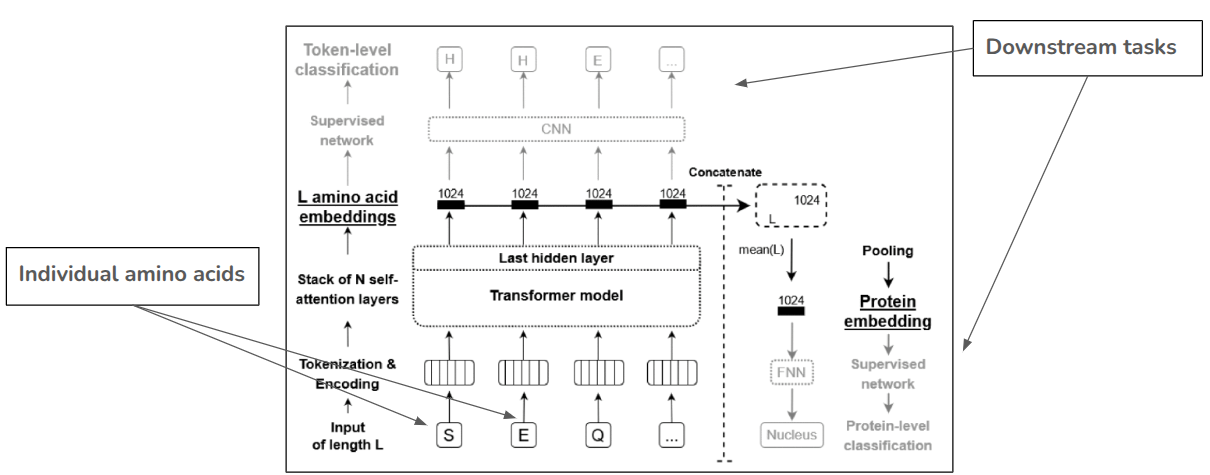
Architecture: Protein Language Model (pLM)
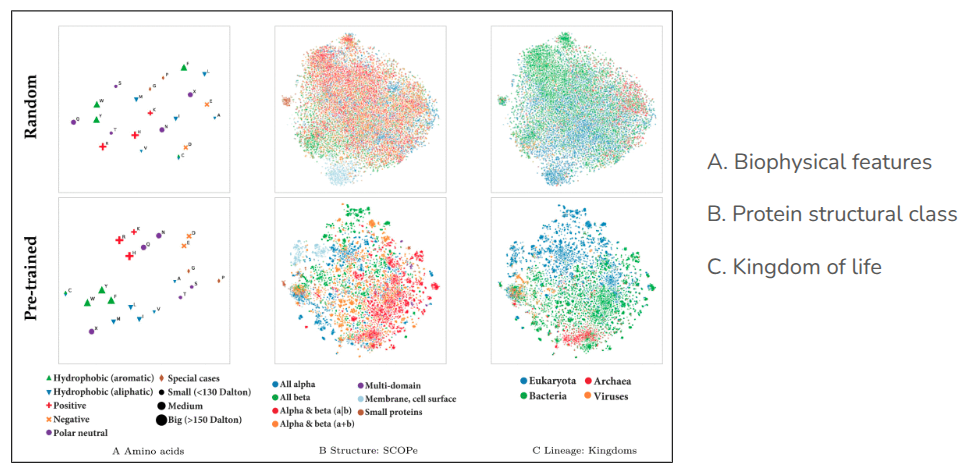
T-SNE embedding projections
Challenges for downstream tasks
- Key tasks
- Fine tuning
- Embedding extraction
- Training challenges
- ProtT5: 1.2 Billion
- Longer training time
- Training or fine-tuning cannot fit on GPU with 15 GB memory (~26 GB)
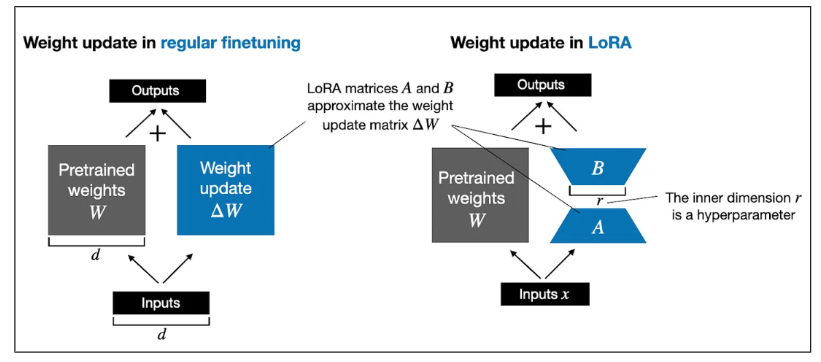
Low-ranking adaption (LoRA)
- Reduce model size
- 1.2 Billion to 3.5 Million parameters
- Fits on GPUs with < 15 GB memory
Use-case: Dephosphorylation (Post-translational modification (PTM)) site prediction
- PTM: chemical modifications to a protein after systhesis
- Crucial for biological processes such as regulating proteins, gene expression, cell cycle, …
- Dephosphorylation
- Removal of a phosphate group from a molecule
- Is less studied and publicly available labeled dataset is small
- Hard to train a large deep learning model
- Fine-tuning might improve site classification accuracy
References
- BERT - https://www.sbert.net/examples/unsupervised_learning/MLM/README.html
- LLM sizes - https://microsoft.github.io/Workshop-Interact-with-OpenAI-models/llms/
- Compute sizes - https://ieeexplore.ieee.org/mediastore/IEEE/content/media/34/9893033/9477085/elnag.t2-3095381-large.gif
- ProtTrans - https://ieeexplore.ieee.org/document/9477085
- LoRA - https://magazine.sebastianraschka.com/p/lora-and-dora-from-scratch
- Dephosphorylation - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8264445/
For additional references, please see tutorial’s References section
- Galaxy Training Materials (training.galaxyproject.org)
Speaker Notes
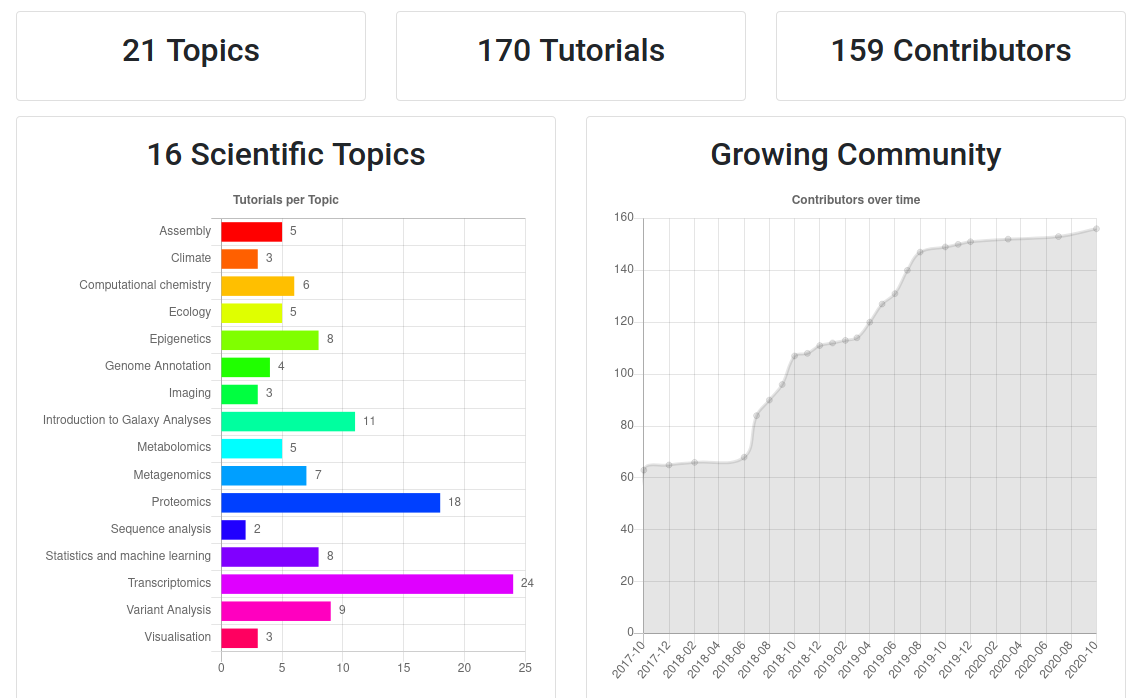
- If you would like to learn more about Galaxy, there are a large number of tutorials available.
- These tutorials cover a wide range of scientific domains.
Getting Help
-
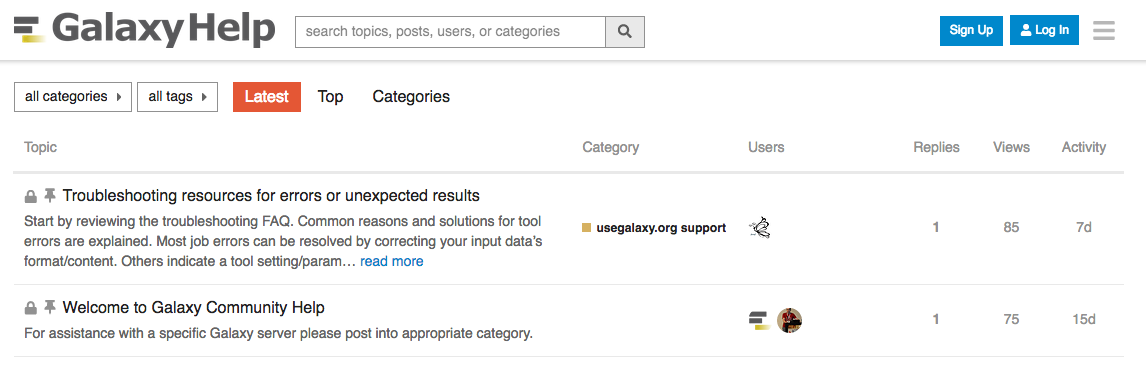
Help Forum (help.galaxyproject.org)
-
Gitter Chat
- Main Chat
- Galaxy Training Chat
- Many more channels (scientific domains, developers, admins)
Speaker Notes
- If you get stuck, there are ways to get help.
- You can ask your questions on the help forum.
- Or you can chat with the community on Gitter.
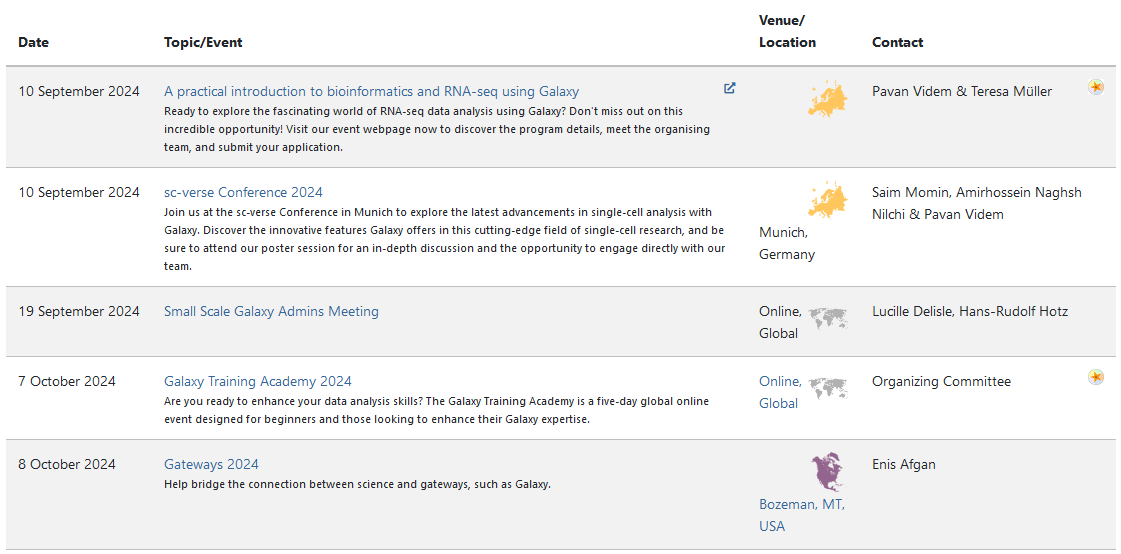
Join an event
- Many Galaxy events across the globe
- Event Horizon: galaxyproject.org/events
Speaker Notes
- There are frequent Galaxy events all around the world.
- You can find upcoming events on the Galaxy Event Horizon.
Key Points
- Training a very large deep learning model from scratch on a large dataset requires exertise and compute power
- Large models such as ProtTrans are trained using millions of protein sequences
- They contain significant knowledge about context in protein sequences
- These models can be used in multiple ways for learning on a new dataset such as fine tuning, embedding extraction, ...
- Fine-tuning using LoRA requires much less time and compute power
- Downstream tasks such as protein sequence classification can be performed using these resources
Thank you!
This material is the result of a collaborative work. Thanks to the Galaxy Training Network and all the contributors! Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.
Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.