name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/introduction" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/introduction/tutorials/options-for-using-galaxy/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" class="cover-logo"/> <br/> <br/> # Options for using Galaxy <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/tnabtaf/" class="contributor-badge contributor-tnabtaf"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/tnabtaf?s=36" alt="Dave Clements avatar" width="36" class="avatar" /> Dave Clements</a> <li> <a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/abretaud?s=36" alt="Anthony Bretaudeau avatar" width="36" class="avatar" /> Anthony Bretaudeau</a> <li> <a href="/training-material/hall-of-fame/pajanne/" class="contributor-badge contributor-pajanne"><img src="https://avatars.githubusercontent.com/pajanne?s=36" alt="Anne Pajon avatar" width="36" class="avatar" /> Anne Pajon</a> <li> <a href="/training-material/hall-of-fame/nsoranzo/" class="contributor-badge contributor-nsoranzo"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/nsoranzo?s=36" alt="Nicola Soranzo avatar" width="36" class="avatar" /> Nicola Soranzo</a> <li> <a href="/training-material/hall-of-fame/alaaelghamry/" class="contributor-badge contributor-alaaelghamry"><img src="https://avatars.githubusercontent.com/alaaelghamry?s=36" alt="Alaa Elghamry avatar" width="36" class="avatar" /> Alaa Elghamry</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00075">gxy.io/GTN:S00075</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="far fa-play-circle" aria-hidden="true"></i><span class="visually-hidden">video-slides</span> <a href="/training-material/videos/watch.html?v=/introduction/tutorials/options-for-using-galaxy/slides">Video slides</a> | <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - Which Galaxy instance should I use? --- # Options for using Galaxy --- ### Options for using Galaxy Galaxy is available in many ways, each with different configurations and capabilities Here are some options ??? Many workshops use Galaxy instances that go away once the workshop is done. This deck lists options for using Galaxy outside of a training context. --- ### usegalaxy.org - [usegalaxy.org](https://usegalaxy.org/) is the Galaxy Project's public server - Hosted at the [Texas Advanced Computing Center (TACC)](https://www.tacc.utexas.edu/) - Lots of - tools - reference genomes - compute power - storage - But also very busy - Averages 250,000 jobs *per month* in 2016 - A single resource cannot scale to meet the demand of the entire world, nor can it support all wanted tools, genomes, ... ??? - usegalaxy.org is the public server, it's hosted at the Texas Advanced Computing Center. - It has lots of tools, reference genomes, computer power, and free storage also. - But it's also very busy, in 2016 it had an average of 250,000 jobs per month, that sometimes could slow down the process - The world of data analysis is huge and full of tools , genomes , and resources - A single resource cannot scale to meet the demand of the entire world --- ### Public Galaxy Servers [.image-25[]](https://galaxyproject.org/public-galaxy-servers/#galaxeast) [.image-25[]](https://galaxyproject.org/public-galaxy-servers/) [.image-25[]](https://galaxyproject.org/public-galaxy-servers/#gigagalaxy) [.image-25[]](https://hyperbrowser.uio.no/hb/) [.image-25[]](https://galaxy.lappsgrid.org/) ??? - There are currently more than 80 public Galaxy servers, and the number keeps growing. - for example, GalaxyEast, an open and powerful galaxy instance for integrative omics data analysis - the Genomic Hyperbrowser to Manage and Analyze collections of genome-wide datasets. - Galaxy Platform Directory which lists platforms where you can use or deploy your own Galaxy Server with minimal effort. --- ### Public Galaxy Servers 1. General purpose genomics servers 1. Domain specific servers<br /> Can be specific to: - *Research methods*: ChIP-Seq, RNA-Seq, repeats, ... - *Organisms*: Pathogens, phage, rice, poplars (!), ... - *Non-genomic domains*: image analysis, social science, natural language processing, ... 1. Tool servers - Host specific tools to make them easy to access and run Complete list: [bit.ly/gxyServers ](https://bit.ly/gxyServers) ??? - There are three types of servers - First is general-purpose genomics servers. - The second is domain-specific servers, which can be specific to: Research methods, Organisms or Non-genomic domains - the third type is Tool servers , the tool servers host specific tools to make them easy to access and run --- ### Semi-public Galaxy services Access based on geography or community membership | Geography | Resource | | ---------- | -------- | | Australia | [Genomics Virtual Lab (GVL)](https://launch.genome.edu.au/launch) | | Canada | [GenAP](https://www.genap.ca/) | | Norway | [Norwegian e-Infrastructure for Life Sciences (NeLS)](https://nels.bioinfo.no/) | | US | [Jetstream](https://galaxyproject.org/cloud/jetstream/) | Complete list: [bit.ly/gxysemipublic ](https://bit.ly/gxysemipublic) ??? - There are some semi-public galaxy servers for which access is restricted according to the user's geographic location or community membership - In Australia there is the Genomics Virtual Lab - In Canada there is GenAP - In Norway there is Norwegian e-Infrastructure for Life Sciences - In the US there is Jetstream --- .pull-left[ ### Public clouds via CloudLaunch .left[[CloudLaunch](https://launch.usegalaxy.org/) currently only supports:] - Jetstream (covered above) - **Amazon Web Services** (AWS) - You need to create an AWS account (which requires a credit card) and then provide your public and secret keys to CloudLaunch ] .pull-right[ [.image-75[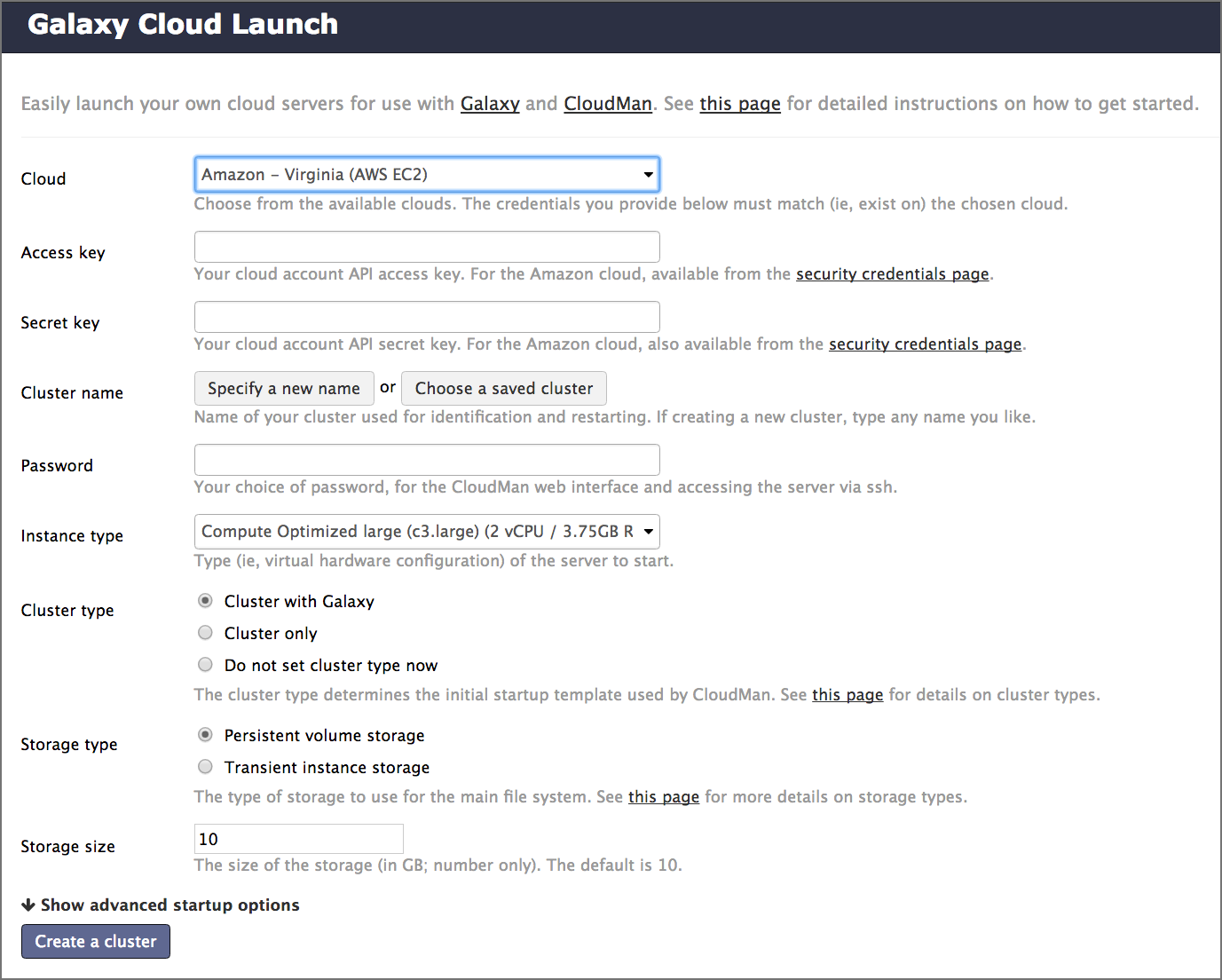]](https://launch.usegalaxy.org/) ] ??? - In addition to servers there is also Public clouds via Galaxy CloudLaunch - Unfortunetly it currently only supports Jetstream and Amazon Web Services - You first need to create an Amazon Web Services account (which requires a credit card) and then provide your public and secret keys to CloudLaunch --- .pull-left[ ### CloudLaunch / CloudMan instances - Comes with 100's of tools and many pre-defined reference genomes - CloudLaunch starts a CloudMan-based Galaxy instance - **CloudMan** is a cloud management tool that moves much / all of the cloud management work into a Graphical user interface ] .pull-right[ [.image-75[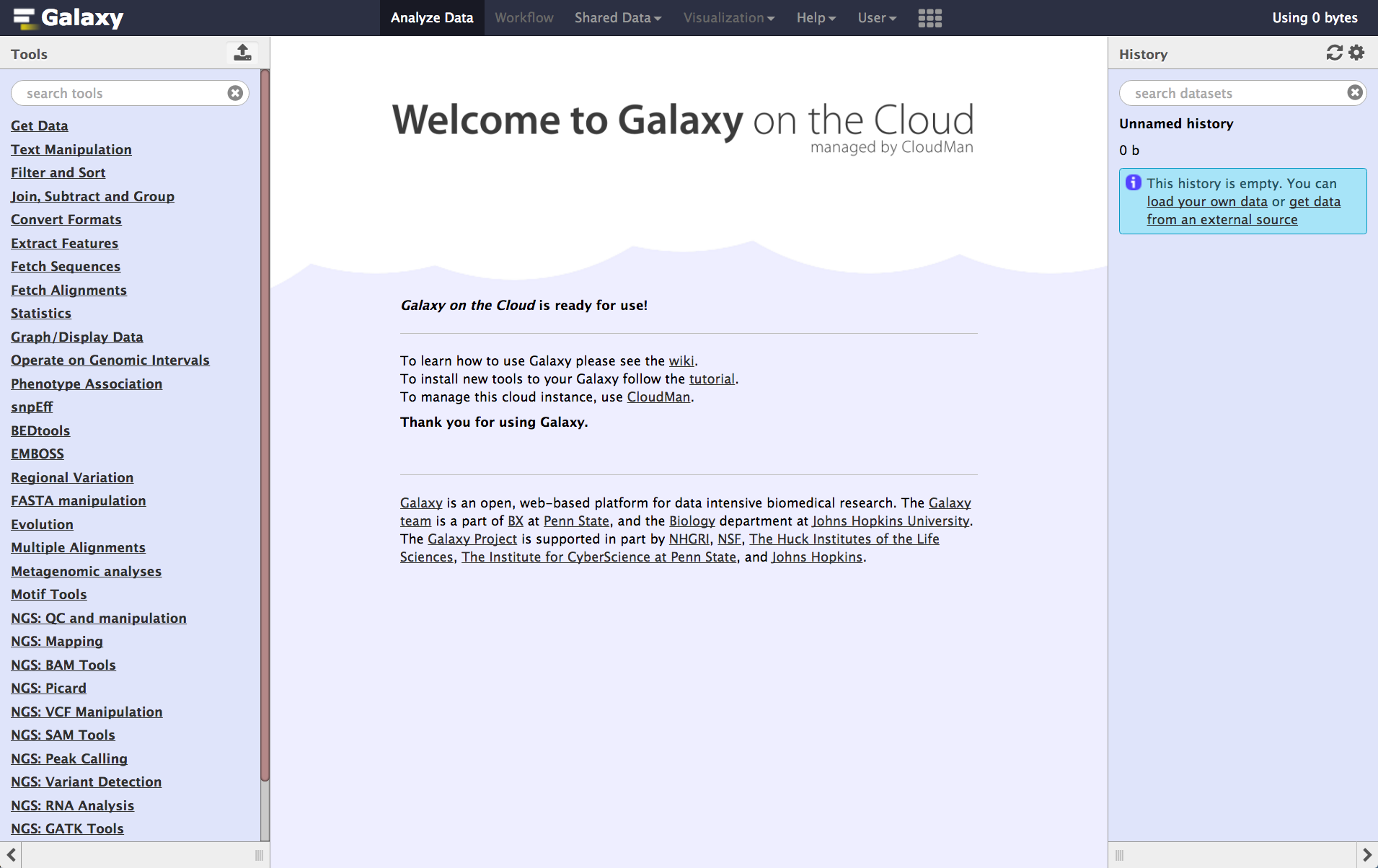]](https://wiki.galaxyproject.org/CloudMan) ] ??? - CloudLaunch Comes with hundredss of tools and many pre-defined reference genomes. - CloudLaunch starts a Galaxy instance called CloudMan. - CloudMan is a cloud management tool that moves so much of the cloud management work into a Graphical user interface. --- ### CloudMan [.image-25[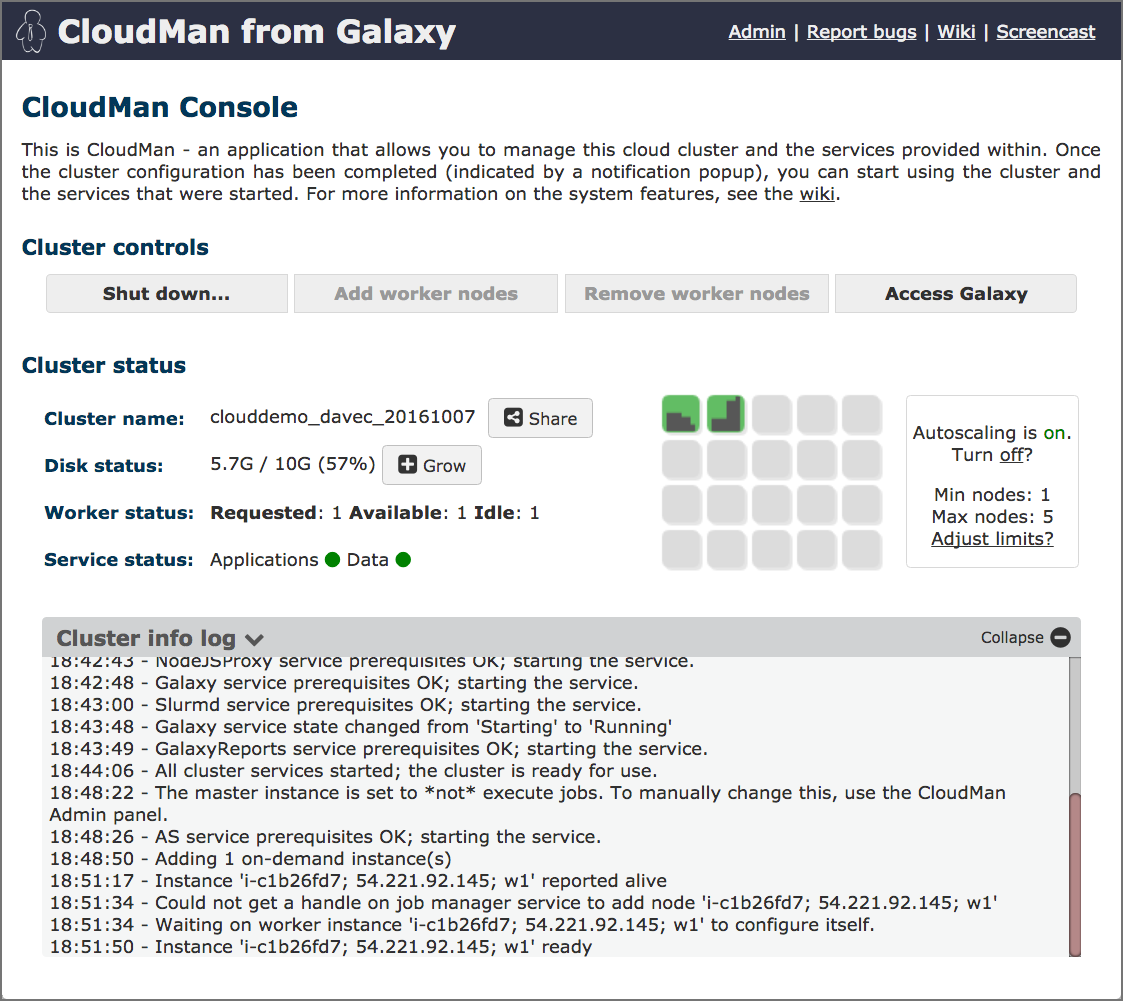]](https://wiki.galaxyproject.org/CloudMan) CloudMan enables you to statically or dynamically scale compute power - This example shows a Galaxy Server with a head node and one worker node, and that is configured to scale up to 5 worker nodes, if demand justifies it - Cloud instances can be incredibly cost effective, but only if you are actually using them when they are up - Paying to have a cloud instance up for two weeks that you only use for 8 hours is not cost effective - **It is very important to shut down and remove everything when you are done** ??? - CloudMan enables you to statically or dynamically scale compute power - Cloud instances can be incredibly cost effective only if you are actually using them when they are up. - you can't have a cloud instance up for two weeks when you only use for 8 hours - It is very important to shut down and remove everything when you are done --- ### Run your own Galaxy locally - Galaxy is [open source software](https://getgalaxy.org/) and can be installed on local compute infrastructure, from lab servers to institutional compute clusters - Installing Galaxy locally is relatively easy, but - the initial install does not include reference genomes and only has a few tools - installing tools and genomes, setting up authentication, and connecting to institutional compute resources all takes work - There are hundreds of local Galaxy installs around the world - Installing tools and genomes has become much easier in recent years, and can now often be done with the Galaxy Admin GUI - Authentication and connecting to institutional compute resources is still heavy lifting ??? - Galaxy is open source software which means it's free to use locally - it can be installed on local compute infrastructure, from lab servers to institutional compute clusters. - Installing Galaxy locally is relatively easy, but you have to consider this - the initial install does not include reference genomes and only has a few tools - installing tools and genomes, setting up authentication, and connecting to institutional compute resources all takes work - installing has become much easier and can now be done with the Galaxy Admin GUI - Authentication and connecting to institutional compute resources is still heavy lifting --- ### Commercial support - You can buy a preconfigured Galaxy server from [BioTeam](https://bioteam.net/) or [NABE<sup>3</sup> International](http://www.nabe-intl.co.jp/seq/takerugalaxy.html) - [Globus Genomics](http://globusgenomics.org/) provides cloud-based Galaxy servers - [Commercially provided consulting and training](https://galaxyproject.org/support/commercial/) are also available ??? - Commercial solutions are also available, for example: - You can buy a preconfigured Galaxy server from BioTeam - Globus Genomics also provides cloud-based Galaxy servers --- ### Which Galaxy instance to use? - Choose based on - size of datasets, available storage, backup - data security - computational requirements - tools installed ??? - In conclusion, choosing which Galaxy instance to use depends on multiple factors like: size of datasets, available storage, backup - you also need to consider data security, computational requirements, and the tools installed --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/tnabtaf/" class="contributor-badge contributor-tnabtaf"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/tnabtaf?s=36" alt="Dave Clements avatar" width="36" class="avatar" /> Dave Clements</a><a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/abretaud?s=36" alt="Anthony Bretaudeau avatar" width="36" class="avatar" /> Anthony Bretaudeau</a><a href="/training-material/hall-of-fame/pajanne/" class="contributor-badge contributor-pajanne"><img src="https://avatars.githubusercontent.com/pajanne?s=36" alt="Anne Pajon avatar" width="36" class="avatar" /> Anne Pajon</a><a href="/training-material/hall-of-fame/nsoranzo/" class="contributor-badge contributor-nsoranzo"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/nsoranzo?s=36" alt="Nicola Soranzo avatar" width="36" class="avatar" /> Nicola Soranzo</a><a href="/training-material/hall-of-fame/alaaelghamry/" class="contributor-badge contributor-alaaelghamry"><img src="https://avatars.githubusercontent.com/alaaelghamry?s=36" alt="Alaa Elghamry avatar" width="36" class="avatar" /> Alaa Elghamry</a> </td> </tr> <tr class="reviewers"> <td><abbr title="These people reviewed this material for accuracy and correctness">Reviewers</abbr></td> <td> <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-badge-small contributor-shiltemann"><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-badge-small contributor-hexylena"><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-badge-small contributor-bgruening"><img src="https://avatars.githubusercontent.com/bgruening?s=36" alt="Björn Grüning avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-badge-small contributor-gallardoalba"><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/nsoranzo/" class="contributor-badge contributor-badge-small contributor-nsoranzo"><img src="https://avatars.githubusercontent.com/nsoranzo?s=36" alt="Nicola Soranzo avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/Alaa-Elghamry/" class="contributor-badge contributor-badge-small contributor-Alaa-Elghamry"><img src="https://avatars.githubusercontent.com/Alaa-Elghamry?s=36" alt="Alaa-Elghamry avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/tnabtaf/" class="contributor-badge contributor-badge-small contributor-tnabtaf"><img src="https://avatars.githubusercontent.com/tnabtaf?s=36" alt="Dave Clements avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/bebatut/" class="contributor-badge contributor-badge-small contributor-bebatut"><img src="https://avatars.githubusercontent.com/bebatut?s=36" alt="Bérénice Batut avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/willdurand/" class="contributor-badge contributor-badge-small contributor-willdurand"><img src="https://avatars.githubusercontent.com/willdurand?s=36" alt="William Durand avatar" width="36" class="avatar" /></a></td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>