name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/admin" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/admin/tutorials/cvmfs/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/gat.png" alt="page logo" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/gat.png" alt="page logo" class="cover-logo" /> <br/> <br/> # Reference Data with CVMFS <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/blankenberg/" class="contributor-badge contributor-blankenberg"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/blankenberg?s=36" alt="Daniel Blankenberg avatar" width="36" class="avatar" /> Daniel Blankenberg</a> <li> <a href="/training-material/hall-of-fame/slugger70/" class="contributor-badge contributor-slugger70"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/slugger70?s=36" alt="Simon Gladman avatar" width="36" class="avatar" /> Simon Gladman</a> <li> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /> Helena Rasche</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00006">gxy.io/GTN:S00006</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="far fa-play-circle" aria-hidden="true"></i><span class="visually-hidden">video-slides</span> <a href="/training-material/videos/watch.html?v=/admin/tutorials/cvmfs/slides">Video slides</a> | <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | <div class="btn-group"> <!-- dropdown with all recordings --> <a href="/training-material/topics/admin/tutorials/cvmfs/recordings/" class="btn btn-default dropdown-toggle topic-icon" data-toggle="dropdown" aria-expanded="false" title="Latest recordings of this material in the GTN Video Library"> <i class="fas fa-video" aria-hidden="true"></i><span class="visually-hidden">video</span> Recordings </a> <ul class="dropdown-menu"> <li><a class="dropdown-item" href="/training-material/topics/admin/tutorials/cvmfs/recordings/index.html#lecture-recording-15-february-2021" title="View the recording for this tutorial"> <i class="fas fa-video" aria-hidden="true"></i><span class="visually-hidden">video</span> Lecture (February 2021) - 3m</a> </li> <li><a class="dropdown-item" href="/training-material/topics/admin/tutorials/cvmfs/recordings/" title="View all recordings for this tutorial"> <i class="fas fa-video" aria-hidden="true"></i><span class="visually-hidden">video</span> View All</a> </li> </ul> </div> </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Have an understanding of what CVMFS is and how it works - Install and configure the CVMFS client on a linux machine and mount the Galaxy reference data repository - Configure your Galaxy to use these reference genomes and indices - Use an ansible playbook for all of the above. --- # Built in Data 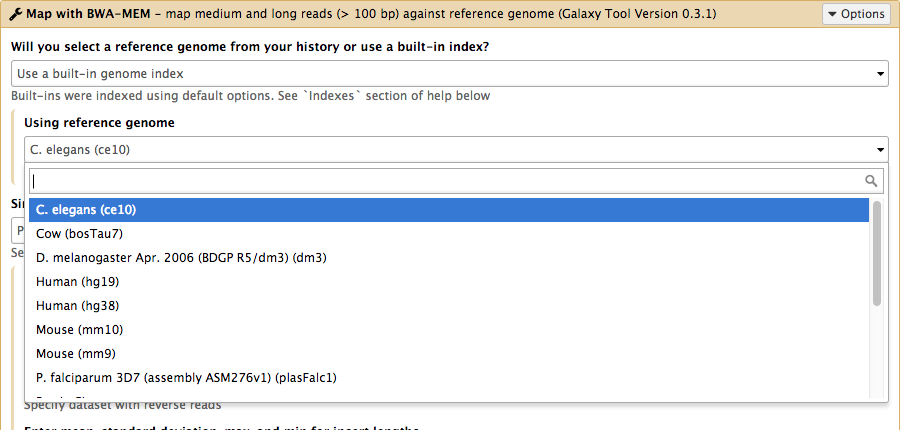 ??? - Many tools need reference data - E.g. this BWA-MEM tool requires a reference genome - But where do you get this data? - And how do tools know where to find it? --- # Data, what data? .large[ * Some genomes are large! Human, Mouse, Coral * Some tools require indices of the genomes. * The indices take a long time to build! * Better to pre-build the indices. ] ??? - Much of this reference data requires calculation to generate it - For example, building dataset indexes - It is better to build them beforehand, so Galaxy has a concept of reference data --- # Data schematics in Galaxy 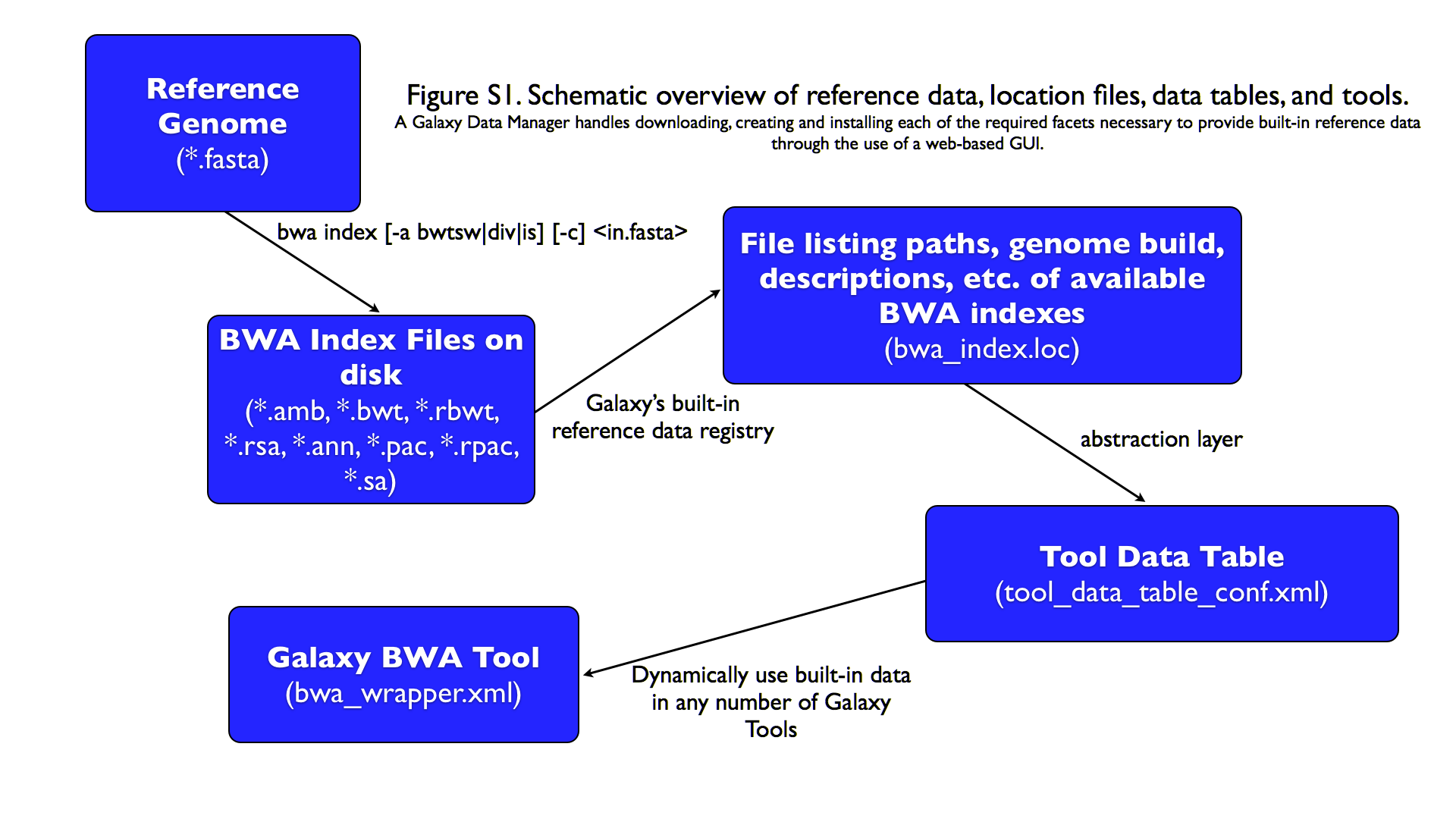 ??? - Here is a more conrete example - A reference dataset is required as input - A tool (e.g. bwa) is used to build the index - The outputs are registered with Galaxy's data registry in a loc file - The tool data table xml file has a listing of all of the loc files - When a user runs the BWA tool, Galaxy knows where to find this reference data --- # Index Generation with Data Manager - Allows for the **creation of built-in** (reference) data - underlying data - data tables - \*.loc files - Specialized Galaxy tools that can only be accessed by an admin - Defined **locally** or installed from **ToolShed** ??? - Data managers are special tools in Galaxy - They create the reference data - And update the loc files --- # "loc" files - Short for location! ```text # #<unique_build_id> <dbkey> <display_name> <file_path> # bosTau7 bosTau7 Cow (bosTau7) /genomes/bosTau7/bwa_mem_index/bosTau7/bosTau7.fa ce10 ce10 C. elegans (ce10) /genomes/ce10/bwa_mem_index/ce10/ce10.fa danRer7 danRer7 Zebrafish (danRer7) /genomes/danRer7/bwa_mem_index/danRer7/danRer7.fa dm3 dm3 D. melanogaster Apr. 2006 (BDGP R5/dm3) (dm3) /genomes/dm3/bwa_mem_index/dm3/dm3.fa hg19 hg19 Human (hg19) /genomes/hg19/bwa_mem_index/hg19/hg19.fa hg38 hg38 Human (hg38) /genomes/hg38/bwa_mem_index/hg38/hg38.fa mm10 mm10 Mouse (mm10) /genomes/mm10/bwa_mem_index/mm10/mm10.fa ``` ??? - Here is an example loc file - They list all of the indexes of a specific type - Some of these indexes will be tool specific like this one, and some will be more general like a list of genomes - This file is updated by the data manager. --- # Where are the data tables? (Usually located in `galaxy/config/tool_data_table_conf.xml`) ```xml <tables> <!-- Locations of indexes in the BWA mapper format --> <table name="bwa_mem_indexes" comment_char="#" allow_duplicate_entries="False"> <columns>value, dbkey, name, path</columns> <file path="tool-data/bwa_index.loc" /> </table> </tables> ``` ??? - The tool data table conf file lists table names, and their associated loc file - Additionally it defines the meaning of each column in the loc file --- # Using reference data in a tool #### bwa.xml ```xml <param name="ref_file" type="select" label="Using reference genome" help="Select genome from the list"> <options from_data_table="bwa_mem_indexes"> <filter type="sort_by" column="2" /> <validator type="no_options" message="No indexes are available" /> </options> <validator type="no_options" message="A built-in reference genome is not available for the build associated with the selected input file"/> </param> ``` ??? - When a tool wants to use the data from those tables, it needs to declare which tables it wants to access (see previous slide) - A select type parameter is created for the user's selections - The tool knows that the options should come from a BWA-MEM indexes loc file - These reference datasets are sorted by column 2, the name - An some validators ensure a helpful message is shown if no data is available --- # Some Problems! - Super complex, right? - Time consuming! - ~30 minutes work just to add a new genome to 1 tool! - Administrator needs to know: - how to index **every** tool - expected format of the reference data - format of the .loc file - Some parts solved by Data Managers - **But there's an easier way!** ??? - This was a lot of information - And very genomics specific in some places - A lot of work to create and update the reference data - But there is a better way! --- # There's a lot of reference data .large[ (and it's hard to keep up with) ] 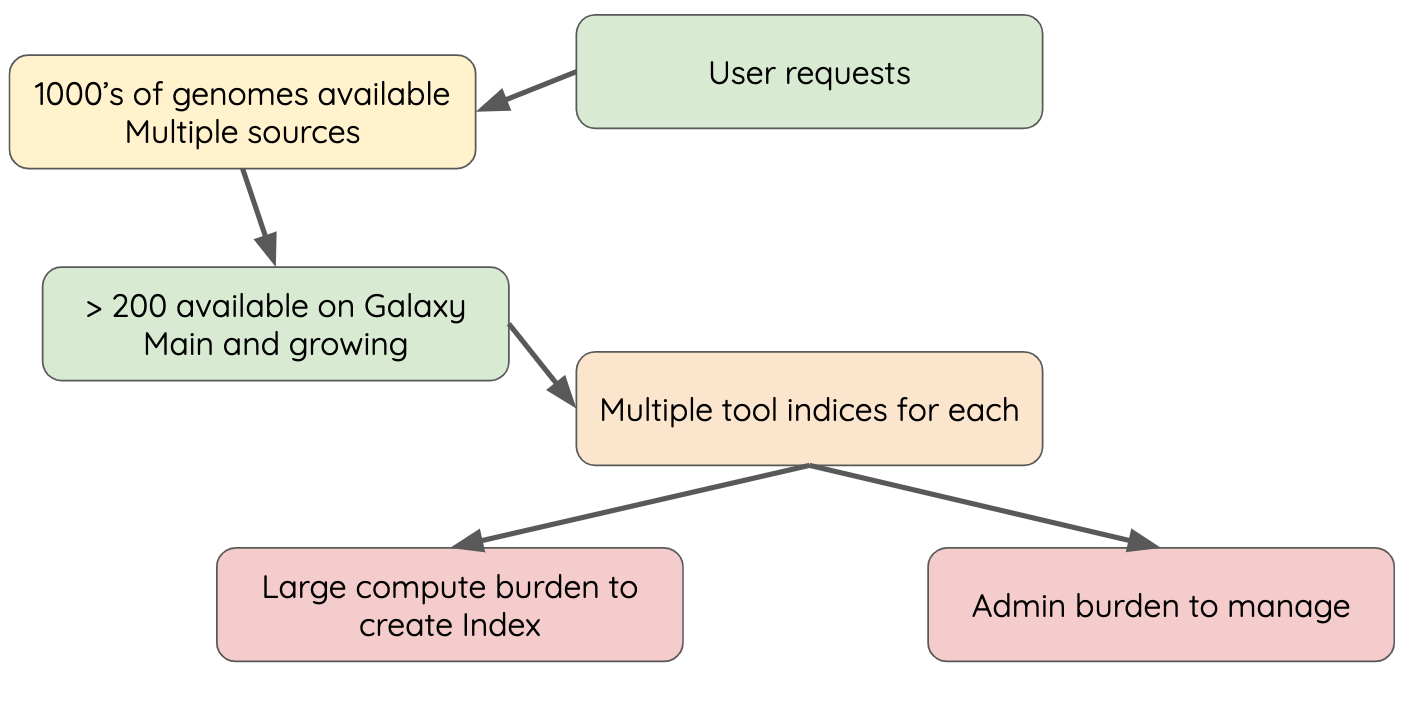 ??? - Imagine going through this process every time a user request comes in - It would be unpleasant --- # CernVM-FS to the rescue - Needed a method of sharing reference data across country efficiently - **CVMFS** is an efficient method for read only data sharing between systems - Originally designed for distributed software installation at Cern - Turns out it's really useful for read only data sets as well - HTTP-based, firewall friendly - All nodes of Galaxy Main get their reference genomes and indices from CVMFS - Shared via mirroring and caching across the country - It's also really useful to share data **globally** - The **usegalaxy.\*** initiative has taken full advantage of this. ??? - CVMFS and the IDC solves these issues - CVMFS provides a global repository of reference data - Originally built by CERN for sharing software, we use it for data - It's an HTTP based protocol and very firewall friendly - All of the usegalaxy.* servers host a CVMFS repository --- # IDC - Group of admins working on the CVMFS data repository - Wish to automate the management of data and generation of new indicies - Come join us: [github.com/galaxyproject/idc](https://github.com/galaxyproject/idc) ??? - The IDC is the other half of the puzzle - CVMFS provides the storage, and the IDC provides the data - Join us on github if you are interested --- # CVMFS Global Structure .widen_image[ 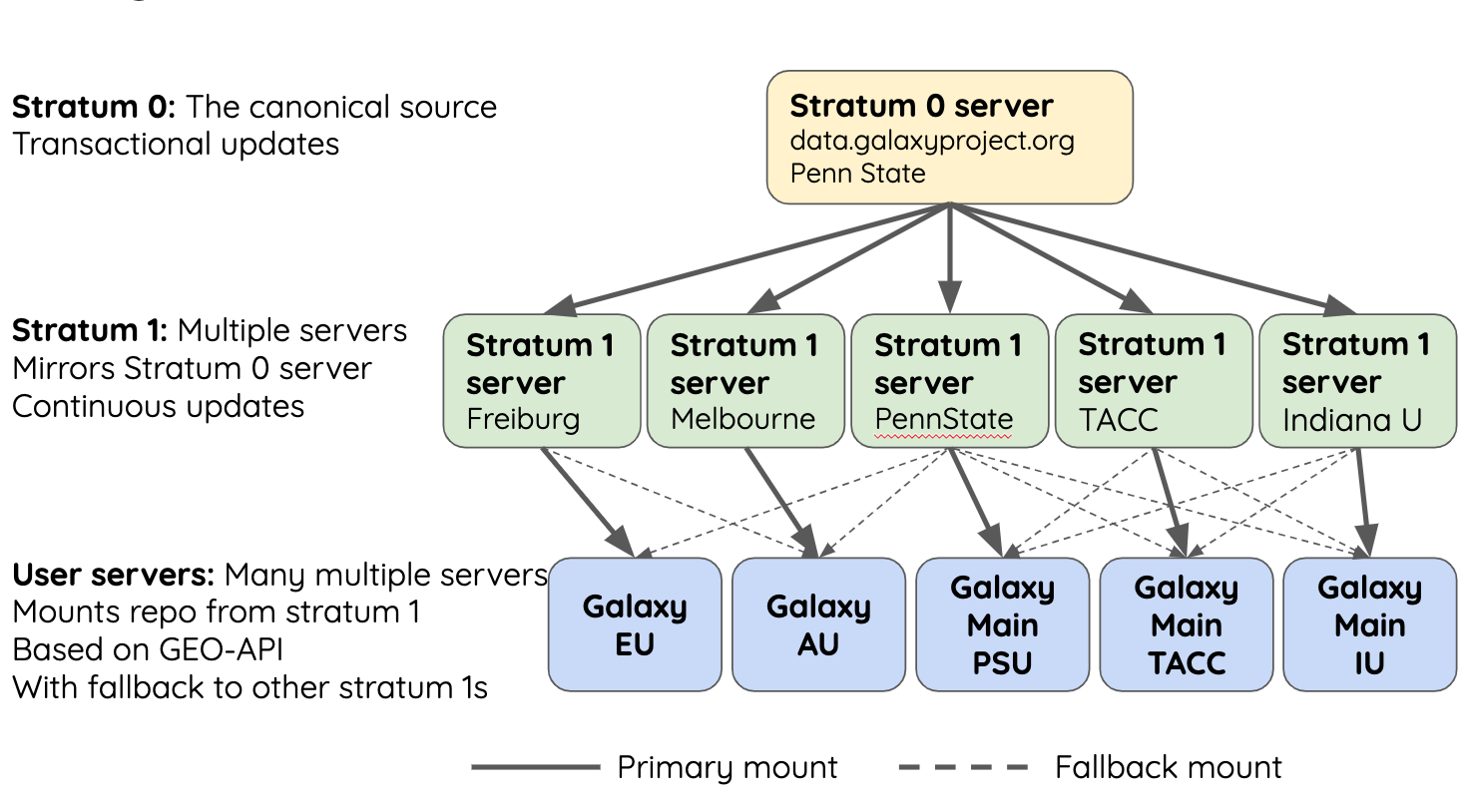 ] ??? - CVMFS has a hierarchical structure - At the top is the stratum 0 server, the original copy of the datasets - This is replicated to the read-only stratum 1 servers - Anyone can connect to these stratum 1 servers - And whenever connections fail, CVMFS will fail over to another mirror - The mirror selection process is based on connection round trip times - As a result, the nearest mirror is usually selected. --- .widen_image[ 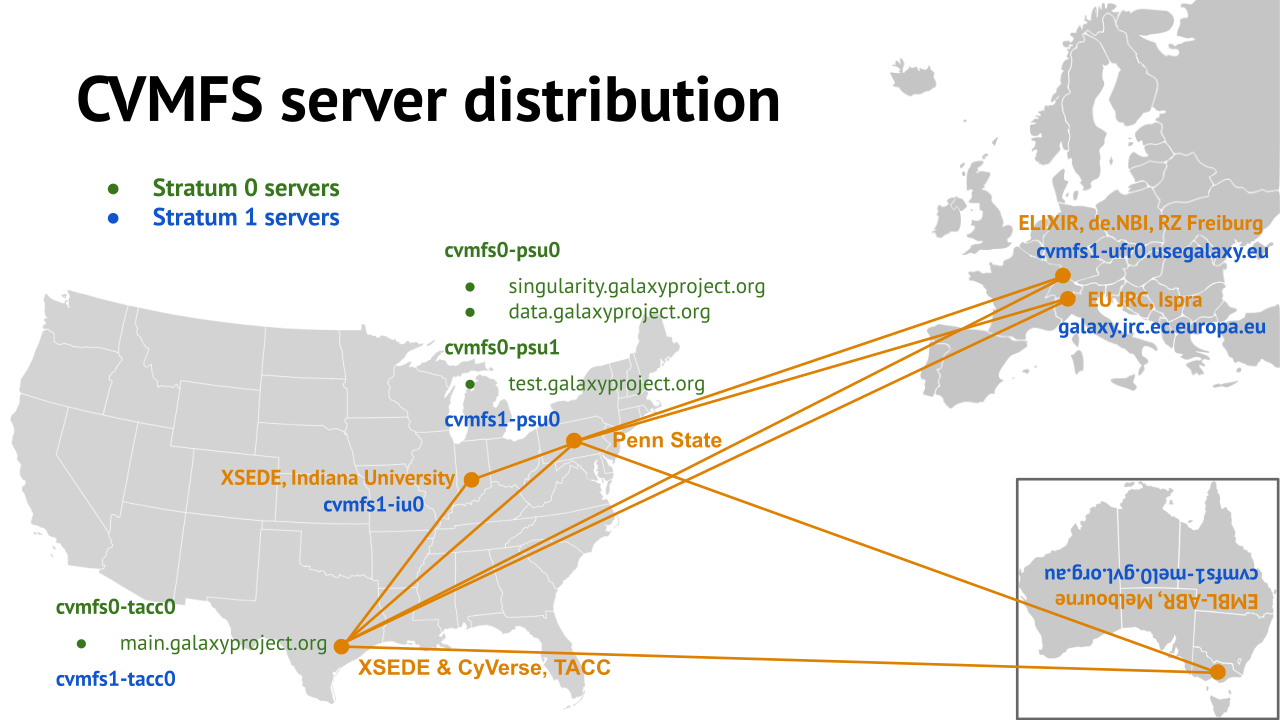 ] ??? - There are CVMFS servers across the entire world - The primary mirrors are run by Galaxy project, Galaxy Europe, and Galaxy Australia - If one of these mirrors fails, you will still be able to use the reference data --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/blankenberg/" class="contributor-badge contributor-blankenberg"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/blankenberg?s=36" alt="Daniel Blankenberg avatar" width="36" class="avatar" /> Daniel Blankenberg</a><a href="/training-material/hall-of-fame/slugger70/" class="contributor-badge contributor-slugger70"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/slugger70?s=36" alt="Simon Gladman avatar" width="36" class="avatar" /> Simon Gladman</a><a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /> Helena Rasche</a> </td> </tr> <tr class="reviewers"> <td><abbr title="These people reviewed this material for accuracy and correctness">Reviewers</abbr></td> <td> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-badge-small contributor-hexylena"><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-badge-small contributor-shiltemann"><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-badge-small contributor-bgruening"><img src="https://avatars.githubusercontent.com/bgruening?s=36" alt="Björn Grüning avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/nsoranzo/" class="contributor-badge contributor-badge-small contributor-nsoranzo"><img src="https://avatars.githubusercontent.com/nsoranzo?s=36" alt="Nicola Soranzo avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/martenson/" class="contributor-badge contributor-badge-small contributor-martenson"><img src="https://avatars.githubusercontent.com/martenson?s=36" alt="Martin Čech avatar" width="36" class="avatar" /></a></td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/gat.png" alt="page logo" style="height: 100px;"/> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>