Introduction to ChIP-Seq data analysis
Contributors
last_modification Published: Jun 12, 2017
last_modification Last Updated: Mar 5, 2024
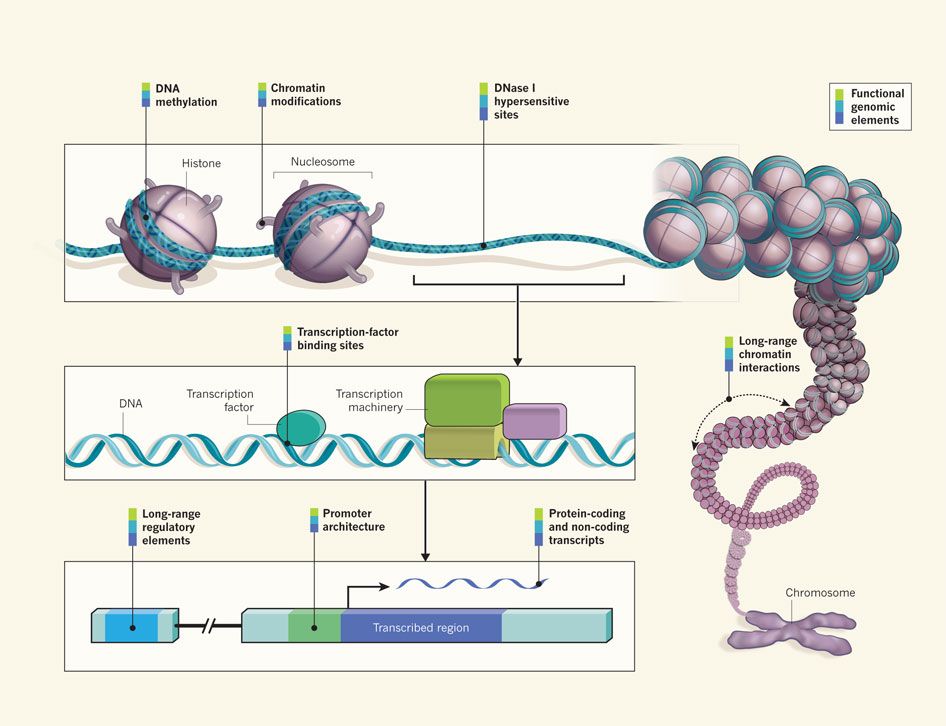
What is ChIP sequencing?
Where my data comes from?
Chromatin immuno-precipitation
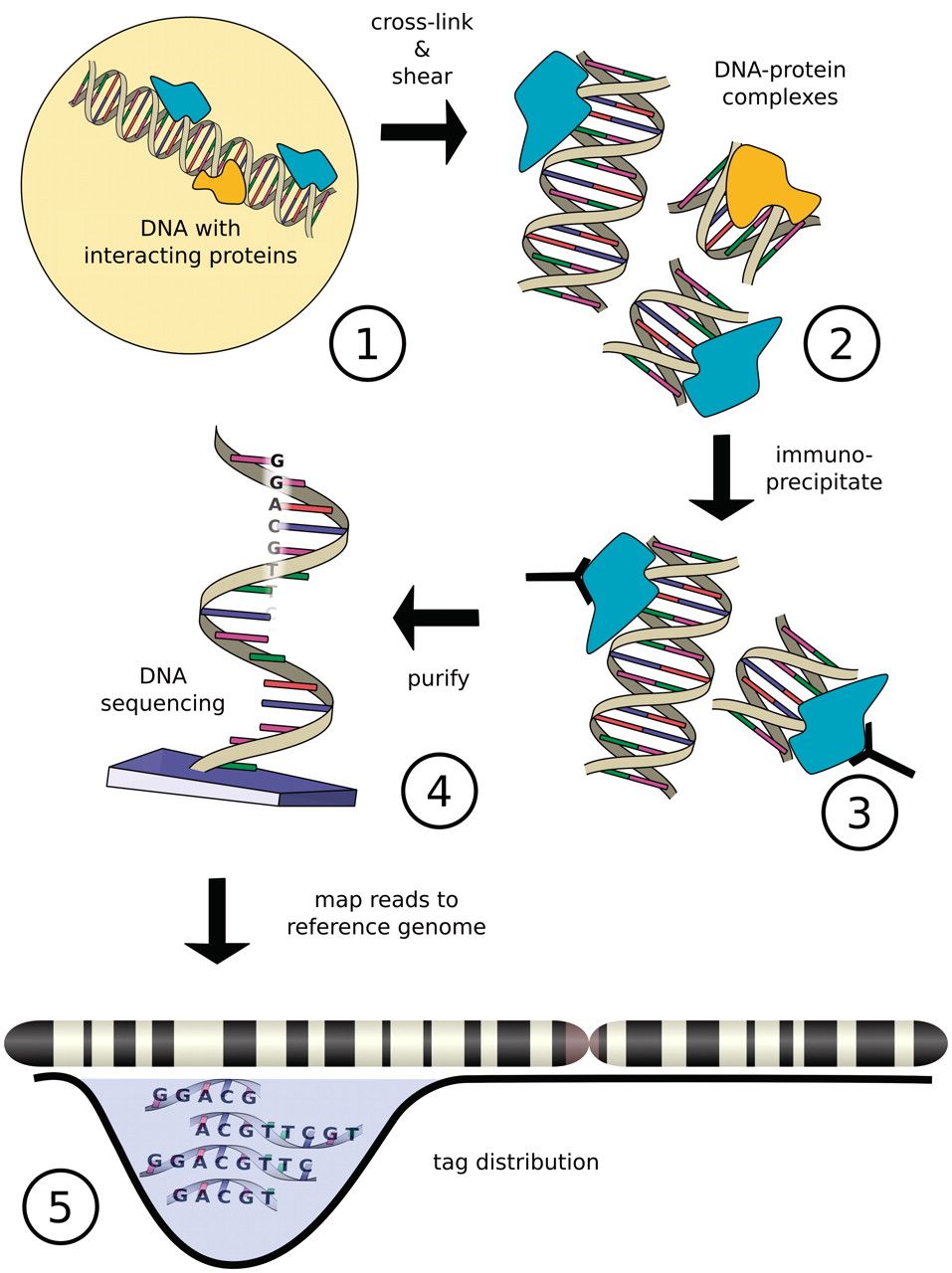
Szalkowski & Schmid, Brief Bioinform, 2011
How to analyze ChIP-seq data?
Common procedures for ChIP-seq data analysis
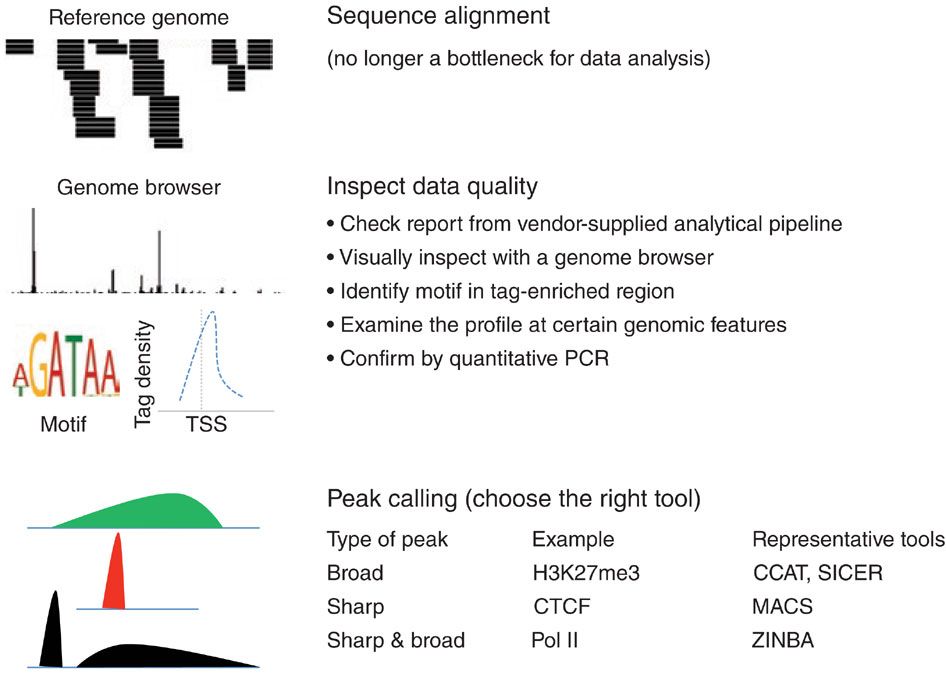
Kidder et al, Nature Immunology, 2011
Thank you!
This material is the result of a collaborative work. Thanks to the Galaxy Training Network and all the contributors! Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.
Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.